- India Today
- Business Today
- Reader’s Digest
- Harper's Bazaar
- Brides Today
- Cosmopolitan
- Aaj Tak Campus
- India Today Hindi

Get 72% off on an annual Print +Digital subscription of India Today Magazine
4500-year-old dna from rakhigarhi reveals evidence that will unsettle hindutva nationalists, the findings of a highly anticipated study of ancient dna from the graveyard of the historic indian town of rakhigarhi reveal evidence that will unsettle many hindutva nationalists.
Listen to Story
Now this somewhat macabre innovation may well resolve one of the most heated debates about the history of India.
As the dust of the petrous bones of a 4,500-year-old skeleton from Rakhigarhi, Haryana, settles, we may have the answer to a few questions that have vexed some of the best minds in history and science -- and a lot of politicians along the way:
Q: Were the people of the Harappan civilisation the original source of the Sanskritic language and culture of Vedic Hinduism? A: No.
Q: Do their genes survive as a significant component in India's current population? A: Most definitely.
Q: Were they closer to popular perceptions of 'Aryans' or of 'Dravidians'? A: Dravidians.
Q: Were they more akin to the South Indians or North Indians of today? A: South Indians.
All loaded questions, of course. A paper suggesting these conclusions is likely to be online in September and later published in the journal Science .
These revelations are part of the long-awaited and much-postponed results of an excavation conducted in 2015 by a team led by Dr Vasant Shinde, an archaeologist and vice chancellor of Pune's Deccan College.
Why did it take so long? One answer was on offer exactly a year ago when this writer spoke to Shinde who was then holding out the promise of publishing the findings in September 2017. "It's a very politically sensitive issue," he said.

The absence of this genetic imprint in the first genome sample of an individual from the Indus Valley culture will bolster what is already a consensus among genetic scientists, historians and philologists: that the Indus Valley culture preceded and was distinct from this population of cattle-herding, horse-rearing, chariot-driving, battle-axe-wielding, proto-Sanskrit-speaking migrants whose ancestry is most evident in high-caste North Indian communities today.

However, he was emphatic in acknowledging that while "a mass movement of Central Asians happened and significantly changed the South Asian genetic make-up", the inhabitants of ancient Rakhigarhi "do not have any affinity with the Central Asians". In other words, while the citizens of the Indus Valley Civilisation had none of this ancestry, you, dear average Indian reader, owe 17.5 per cent of your male lineage to people from the Steppe.
It's worth noting that this genetic footprint is of an entirely more impressive order than the relatively inconsequential biological legacy of Islamic or European colonial invasions that often preoccupy the political imagination in India.
So much for what we have now learned about who our 4,500-year-old ancestor 'I4411' was not. What about who he was? The short answer, says Rai, is that I4411 "has more affinity with South Indian tribal populations". Notably, the Irula in the Nilgiri highlands.
A draft of the paper argues that this individual could be modelled as part of a clade [a group sharing descent from a common ancestor] with the Irula but not with groups with higher proportions of West Eurasian related ancestry such as Punjabis, and goes on to suggest that the inhabitants of Rakhigarhi probably spoke an early Dravidian language.
Most mainstream historians have discarded the 'Aryan invasion theory' or 'AIT' as an oversimplification
However, the results also show clear evidence of mixing with another population from outside the subcontinent, labelled 'Iranian agriculturalist'. This is a population that had been identified in earlier studies of ancient DNA and is consistent with the hypothesis that some agricultural technologies were introduced to the subcontinent through contact with the 'fertile crescent' in West Asia, widely regarded as one of the birthplaces of Eurasian agriculture in the 5th-8th millennium BC.
For an older generation of Indians, the Rakhigarhi results may sound like a reboot of half-remembered schoolbooks: 'Dravidian' Harappans followed by Vedic horsemen from the Steppe. And for anyone who has been following more recent developments in population genetics too, the latest findings will sound familiar.
Meanwhile, in the popular press, coverage of recent discoveries in the archaeology or genetics of Harappan India has been obsessively and distractingly focused on the 'Aryan invasion theory'. What gives? And why does it matter? The answer has to do with the fact that recent years have been a very busy time in ancient Indian history. And modern Indian politics.
SKULDUGGERY
In the months preceding the news of the Rakhigarhi findings, anticipation was high, and fuelled by a series of related research papers and their journalistic glosses, an amusing if acrimonious debate erupted in the social media and the blogosphere. Shinde for his part was given to dropping broad hints that the Rakhigarhi results would point to a 'continuity' between the population of the ancient town and its present-day inhabitants (predominantly Jats, a population marked by pronounced R1a Steppe ancestry).
Perhaps it should be no surprise, in these fractious times, that fake news would be deployed as a weapon in the civil war that has consumed ancient Indian history. In January this year, a Hindi newspaper carried an article purportedly based on an interview with Rai, asserting that the Rakhigarhi DNA was, in fact, a close match for North Indian Brahmins and that the findings would establish that India was the 'native place' of the Indo-European language family.
"Utter crud!" was the reaction of David Wesolowski, host of the Eurogenes blog-well regarded by some of the world's leading geneticists as a go-to site for the latest debate. Wesolowski's site witnessed frequent arguments over the likelihood that Rakhigarhi DNA would turn up the R1a1 marker.
Here, extended and nuanced discussions of the finer points of molecular evidence would often conclude with kiss-offs along the lines of "you're an idiot" or "you're going to need psychiatric help when the results are out". In the event, Wesolowski's own prediction, "Expect no R1a in Harappa but a lot of ASI [Ancestral South Indian]", would prove to be spot on.
The single most startling revelation of the Rakhigarhi research may be the complete absence of any reference to the genetic marker R1a1, often loosely called 'the 'Aryan gene'
Behind the surly invective and the journalistic misdirection were rumours and whispers of a face-off between a rising tide of scientific evidence and the political pressures of nativist, Hindutva sentiments.
The saga of 'Hindutvist history' is by now another familiar tale, with its origins in early Hindu nationalist reaction to colonial archaeology and linguistics, a monomaniacal obsession with refuting the 'Aryan invasion theory'.
It is perhaps most clearly expressed in an irate passage from former RSS sarsanghchalak M.S. Golwalkar's screed Bunch of Thoughts (1966): "It was the wily foreigner, the Britisher, who carried on the insidious propaganda that we were never one nation, that we were never the children of the soil but mere upstarts having no better claim than the foreign hordes of Muslims or the British over this country."
In recent years, this resentful impulse has focused particularly intently on asserting the wishful conclusion that the Indus Valley Civilisation itself must be 'Vedic'. This has understandably gained traction in the popular imagination in tandem with the political rise of Hindutva. In 2013, Amish Tripathi, a bestselling author of 'Hinduistical fantasy' novels, gave vent to the keening desire for a 'Vedic IVC' in a short fiction in which future archaeologists discover clinching evidence "that the Indus Valley Civilisation and the Vedic-erroneously called Aryan-civilisation were one and the same." The story is poignantly titled, 'Science Validates Vedic History'.
Inevitably, the advent of a BJP majority government in the general elections of 2014 has given new energy -- and funding-to the self-gratifying urges of Hindutvist history.

In March this year a Reuters report revealed details of a meeting of a 'history committee' convened by Sharma at the office of the Director General of the Archaeological Survey of India in January 2017. Its task, according to the committee chairman K.N. Dixit, was "to present a report that will help the government rewrite certain aspects of ancient history".
The minutes of the meeting apparently "set out its aims: to use evidence such as archaeological finds and DNA to prove that today's Hindus are directly descended from the land's first inhabitants many thousands of years ago, and make the case that ancient Hindu scriptures are fact, not myth".
Yet, if the 'rewriting of Indian history' was lurching ahead on the Hindutva fringe of academia, mainstream science was steadily advancing in quite another direction.
In March this year, the Harvard population geneticist David Reich published an overview of the state of research in his field, the surprise bestseller Who We Are and How We Got Here , including an account of how the extreme sensitivity of leading Indian scientists about earlier evidence suggesting an ancient migration of Eurasian people from the Northwest into the subcontinent had nearly scuppered an important scientific collaboration in 2008.

But the same dynamic appears to have emerged this year around a paper involving both Reich and his team at Harvard on the one hand and the scientists leading the Rakhigarhi project on the other. Entitled, rather flatly, The Genomic Formation of South and Central Asia, this paper (usually referred to by the shorthand 'M Narasimhan et al') -- made public as a 'pre print' in April -- would make headlines in the Indian press and social media and reveal some more of the political pressures that colour research on ancient Indian history today.
Shinde said that he had complained to Reich about an earlier draft of that paper, and insisted that any reference to 'migrations' into South Asia be avoided. Or else. He suggested the more ambivalent term 'interaction' be used instead.
As the results of the Rakhigarhi study leak steadily into the public domain, a political backlash seems inevitable
Given that Shinde controlled access to the Rakhigarhi samples which Reich was keen to work on, this would have been a potent threat, and indeed the paper manages to eschew the term 'migration' entirely while ultimately making more potent statements about the impact of post-Harappan 'Middle to Late Bronze Age' (MLBA) Steppe populations on the Indian gene pool.
However, the timing of the paper remains curious to say the least, given that it would have benefitted from the Rakhigarhi data which it seemed to pre-empt -- despite the fact that several of its co-authors, including Rai, Shinde, Thangaraj, Narasimhan and Reich now share credit for the mysteriously delayed paper.
The official word on this was that the Rakhigarhi research was behind schedule due to the 'contamination of one sample', but at the time the geneticist community was abuzz with rumours that the slowdown was because of the Indian team's discomfort with politically inconvenient results.
According to one US-based researcher, who prefers to remain anonymous, "It was common knowledge through the grapevine that the Harvard team became impatient and eventually pushed to release their preprint before Indian colleagues were totally comfortable. Some samples [read 'Rakhigarhi'] were removed because of disagreements between collaborators."

However, Shinde is no geneticist, and from what we now know, the Rakhigarhi study endorses the findings of the Narasimhan paper -- indeed, it can be seen as a companion piece to that earlier work of the common authors.
Significantly, while Narasimhan and others predicted a model of the Harappan genome using samples of DNA from ancient skeletons of apparent Indus Valley 'visitors' found in sites that were in trading contact with the Harappans, as well as remains of post-Harappan (1200-BC-1 CE) individuals from Swat, the Rakhigarhi paper suggests that this model was accurate. It recommends that the Narasimhan paper's tentative label of 'Indus Valley periphery' for this model is a significant match for I4411 of Rakhigarhi and this genetic cluster should now be recognised as the 'Harappan cline'.
IT'S STILL COMPLICATED
As the results of the Rakhigarhi study leak steadily into the public domain, a political backlash seems inevitable -- and largely predictable: some exultation from Dravidianists and the legion of anti-Hindutva Indians for many of whom the fall of Delhi in the 2014 election is seen as a calamitous replay of that fabled 'Vedic Aryan invasion'.
And we can expect sullen scepticism from the saffron right. Intriguingly, some of the strongest reservations about the Rakhigarhi project have already been expressed from an unexpected quarter: established historians.
Romila Thapar, always a name to reckon with in ancient Indian history and a perennial target of Hindutva polemic, has followed the genetics story keenly, but expressed her reservations about this new science.
As it turns out, the Rakhigarhi research was not without glitches -- apparently, a misleading 'East Asian' signal in the early data is the reason why the Korean scientists who first worked on the samples may not be credited in the final paper.
Meanwhile, another respected historian, Nayanjot Lahiri, declared complete disinterest in the work on 'Harappan DNA', voicing impatience at the obsession with the 'Aryan' question and scepticism about the narrow sampling of ancient genetic material. "As far as the whole question of Aryans and the Vedic component in the Indus Valley Civilisation goes, until the Harappan script is deciphered, it's not decided," she says.

Similarly, any impulse to equate the apparent Dravidian affinities of ancient Indus Valley people with the culture and people of South India today or to cast the latter as the 'original inhabitants' of the subcontinent would be an exaggeration.
India is composed of a large number of small populations
Quite apart from the fact that the people and cultures across the subcontinent today display evidence of having mixed with each other (and populations beyond the borders of present day India) over millennia, there is also no population in the region that can claim to represent a 'pure' lineage of ancient Indians.
Not even the Irula or any other South Indian or 'Adivasi' group. Nor should the evidence of the deeply intertwined genetic history of Indian communities lull anyone into a cosy fable of Indic cosmopolitanism.
What our DNA tells us instead is that while India witnessed phases of extensive genetic mixing for a millennium after the collapse of the Indus Valley Civilisation, this was followed by a long period of deep endogamy -- which has been a uniquely unhealthy stamp of the subcontinent.
Reich summed it up in his recent book: "People tend to think of India, with its more than 1.3 billion people, as having a tremendously large population. But genetically, this is an incorrect way to view the situation. The Han Chinese are truly a large population. They have been mixing freely for thousands of years The truth is that India is composed of a large number of small populations."

At the end of the day, Nehru's vision too is a modern nationalist fantasy. In the years to come, we are certain to discover much more about the enduringly mysterious civilisation of the Harappans and what elements of culture and social behaviour they bequeathed us -- along with their genes. For now, miraculously, their ears are speaking. We would do well to listen for a while.
READ | 3 strands of ancestry
Rare Ancient DNA Provides Window Into a 5,000-Year-Old South Asian Civilization
The Indus Valley Civilization flourished alongside Mesopotamia and Egypt, but the early society remains shrouded in mystery
Brian Handwerk
Science Correspondent
/https://tf-cmsv2-smithsonianmag-media.s3.amazonaws.com/filer/df/8a/df8a7647-d6e7-4f4b-ab4b-ab3046ddafa8/210155.jpg)
During the last few millennia B.C., beginning roughly 5,000 years ago, great civilizations prospered across Eurasia and North Africa. The ancient societies of Mesopotamia and Sumer in the Middle East were among the first to introduce written history; the Old, Middle and New Kingdoms of Egypt established complex religious and social structures; and the Xia, Shang and Zhou dynasties ruled over ever advancing communities and technologies in China. But another, little understood civilization prevailed along the basins of the Indus River, stretching across much of modern Afghanistan and Pakistan and into the northwestern regions of India.
This Indus Valley Civilization (IVC), also called the Harappan civilization after an archaeological site in Pakistan, has remained veiled in mystery largely due to the fact that scholars have yet to make sense of the Harappan language , comprised of fragmented symbols, drawings and other writings. Archaeological evidence gives researchers some sense of the daily lives of the Harappan people, but scientists have struggled to piece together evidence from ancient DNA in the IVC due to the deterioration of genetic material in the hot and humid region—until now.
For the first time, scientists have sequenced the genome of a person from the Harappan or Indus Valley Civilization, which peaked in today’s India-Pakistan border region around 2600 to 1900 B.C. A trace amount of DNA from a woman in a 4,500-year-old burial site, painstakingly recovered from ancient skeletal remains, gives researchers a window into one of the oldest civilizations in the world. The work, along with a comprehensive analysis of ancient DNA across the Eurasian continent, also raises new questions about the origins of agriculture in South Asia.
The ancient Harappan genome, sequenced and described in the journal Cell , was compared to the DNA of modern South Asians, revealing that the people of the IVC were the primary ancestors of most living Indians. Both modern South Asian DNA and the Harappan genome have a telltale mixture of ancient Iranian DNA and a smattering of Southeast Asian hunter-gatherer lineages. "Ancestry like that in the IVC individuals is the primary ancestry source in South Asia today,” co-author David Reich, a geneticist at Harvard Medical School, said in a statement. “This finding ties people in South Asia today directly to the Indus Valley Civilization.”

The genome also holds some surprises. Genetic relationships to Steppe pastoralists, who ranged across the vast Eurasian grasslands from contemporary Eastern Europe to Mongolia, are ubiquitous among living South Asians as well as Europeans and other people across the continent. But Steppe pastoralist DNA is absent in the ancient Indus Valley individual, suggesting similarities between these nomadic herders and modern populations arose from migrations after the IVC’s decline.
These findings influence theories about how and when Indo-European languages spread widely across the ancient world. And while shared ancestry between modern South Asians and early Iranian farmers has fueled ideas that agriculture arrived in the Indo-Pakistani region via migration from the Fertile Crescent of the Middle East, the ancient Harappan genes show little contribution from that lineage, suggesting that farming spread through an exchange of ideas rather than a mass migration, or perhaps even arose independently in South Asia.
“The archaeology and linguistic work that had been carried out for decades was really at the forefront of our process,” says Vagheesh Narasimhan , a Harvard University genomicist and co-author of the new study. “These projects bring a new line of genetic evidence to the process, to try to show the impact that the movement of people may have had as part of these two great cultural transformations of agriculture and language.”
The large, well-planned cities of the IVC included sewer and water systems, as well as long-distance trade networks that stretched as far as Mesopotamia. But despite its former glory, the civilization was unknown to modern researchers until 1921, when excavations at Harappa began to uncover an ancient city. The Harappans have remained something of a mystery ever since, leaving behind extensive urban ruins and a mysterious language of symbols and drawings, but few additional clues to their identity. What ultimately befell the Harappan civilization is also unclear, though a changing climate has been posited as part of its downfall.
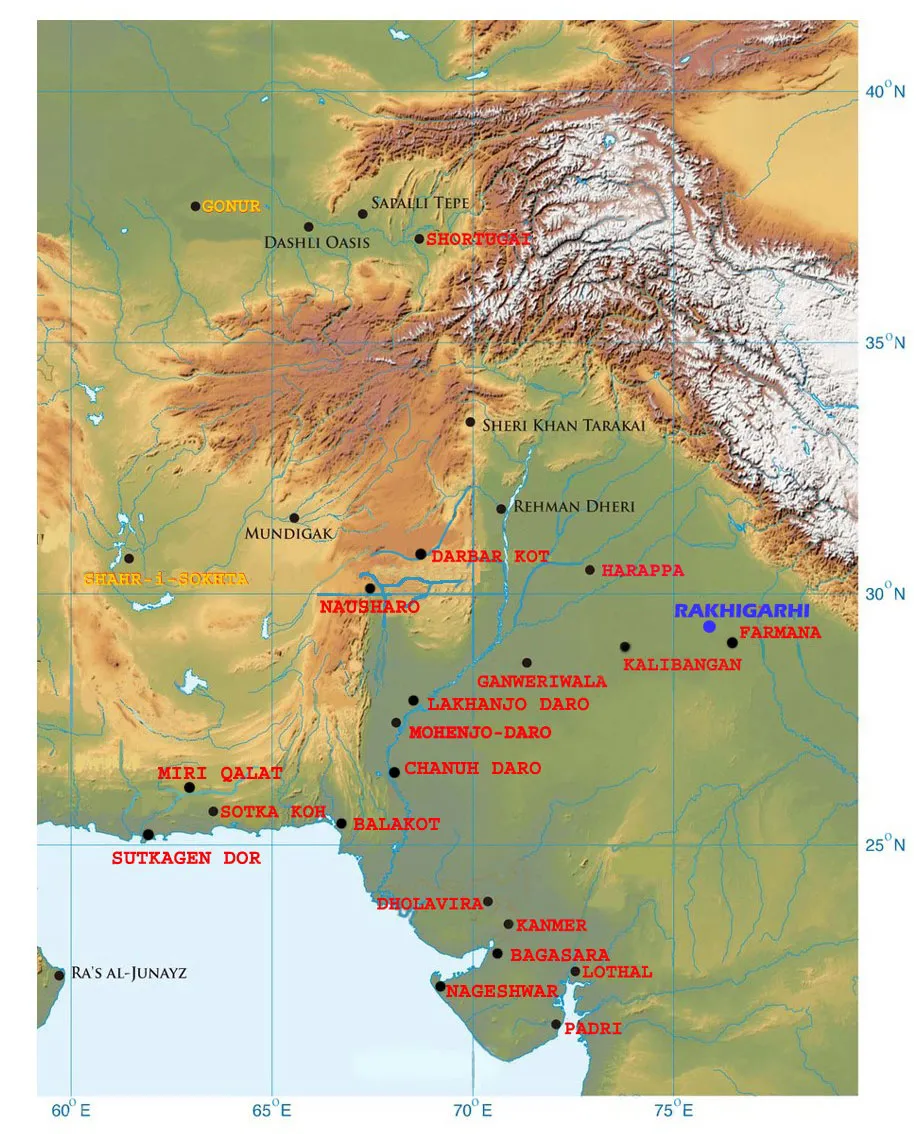
Scientists have a notoriously difficult time recovering ancient DNA in South Asia, where the subtropical climate typically makes genetic preservation impossible. It took a massive, time-consuming effort to produce the genome from remains found in the cemetery at Rakhigarhi, the Harappans’ largest city, located in the modern Indian state of Haryana. Scientists collected powder from 61 skeletal samples, but just one contained a minute amount of ancient DNA. That sample was sequenced as much as possible, generating 100 different collections of DNA fragments, called libraries, each of which were too incomplete to yield their own analysis.
“We had to pool 100 libraries together and sort of hold our breath, but we were fortunate that that yielded enough DNA to then do high resolution population genetics analysis,” Narasimhan says. “I think if anything, this paper is a technical success story,” he adds, noting that the approach holds promise for sourcing DNA in other challenging locales.
A single sample is not representative of a widespread population that once included a million or more people, but a related study published today in Science lends some wider regional context. Several of the same authors, including Narasimhan and Reich, and dozens of international collaborators, authored the largest ancient DNA study published to date. Among the genetic sequences from 523 ancient humans are individuals from sites as far flung as the Eurasian Steppe, eastern Iran and Iron Age Swat Valley in modern Pakistan.
The team found that among many genetically similar individuals, a handful of outliers existed who had ancestry types completely different from those found around them.
Eleven such individuals found at sites in Iran and Turkmenistan were likely involved in interchange with the Harappan civilization. In fact, some of these outlier individuals were buried with artifacts culturally affiliated with South Asia, strengthening the case that they were connected to the IVC.
“This made us hypothesize that these samples were migrants, possibly even first-generation migrants from South Asia,” Narasimhan says. The IVC genome from Rakhigarhi shows strong genetic similarities to the 11 genetic outliers in the large study of ancient humans, supporting the idea that these individuals ventured from the Harappan civilization to the Middle East. “Now we believe that these 12 samples, taken together, broadly represent the ancestry that was present in [South Asia] at that time.”
The first evidence of agriculture comes from the Fertile Crescent, dating to as early as 9,500 B.C., and many archaeologists have long believed that the practice of growing crops was brought to South Asia from the Middle East by migrants. Earlier DNA studies seemed to bear out this idea, since South Asians today have significant Iranian ancestry.
“I really found their analysis to be very exciting, where they look at ancient DNA samples from different time scales in Iran and try to correlate how the Iranian ancestry in South Asians is related to those different groups,” says Priya Moorjani, a population geneticist at UC Berkeley not involved in the Cell study of the IVC genome.
However, the new analysis shows that the first farmers of the Fertile Crescent appear to have contributed little, genetically, to South Asian populations. “Yet similar practices of farming are present in South Asia by about 8,000 B.C. or so,” says Moorjani, a co-author on the wider population study of South and Central Asia. “As we are getting more ancient DNA, we can start to build a more detailed picture of how farming spread across the world. We’re learning, as with everything else, that things are very complex.”
If farming did spread from the Fertile Crescent to modern India, it likely spread via the exchange of ideas and knowledge—a cultural transfer rather than a significant migration of western Iranian farmers themselves. Alternatively, farming could have arisen independently in South Asia, as agricultural practices started to sprout up in many places across Eurasia during this time.
Ancient IVC ancestry holds other mysteries as well. This civilization was the largest source population for modern South Asians, and for Iron Age South Asians as well, but it lacks the Steppe pastoralist lineages common in later eras. “Just like in Europe, where Steppe pastoralist ancestry doesn’t arrive until the Bronze Age, this is also the case in South Asia,” Narasimhan says. “So this evidence provides information about the timing of arrival of this ancestry type, and their movement parallels the linguistic phylogeny of Indo-European languages, which today are spoken in places as far away as Ireland to New Delhi.”
The authors suggest Indo-European languages may have reached South Asia via Central Asia and Eastern Europe during the first half of the 1000s B.C., a theory evidenced by some genetic studies and similarities between Indo-Iranian and Balto-Slavic languages.
Narasimhan hopes that more genetic data can help clear up this ancient puzzle—especially by exploring where DNA dovetails or differs with findings from other lines of evidence.
“We’re trying to look at when and how archaeological cultures are associated with a particular genetic ancestry, and whether there’s any linguistic connections,” he says. “To understand human history, you really need to integrate these three lines.”
Get the latest Science stories in your inbox.
Brian Handwerk | READ MORE
Brian Handwerk is a science correspondent based in Amherst, New Hampshire.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Sign up for alerts
- 04 September 2019
Where did the Indus Valley people come from?
You can also search for this author in PubMed Google Scholar

Skeleton of the Rakhigarhi woman, whose genome the scientists analysed. © Vasant Shinde/DCPGRI
The first genome sequence of a woman from Rakhigarhi, the largest town in Indus Valley Civilisation, has shed some light on the origins of people who built this ancient civilisation around five millennia ago 1 .
The DNA belongs to a woman who was buried four to five millennia ago in Rakhigarhi, now part of Haryana in India. Her genes point to an ancestry of ancient Iranians and Southeast Asian hunter-gatherers.
Genetic scientists say this does not mean that her ancestors lived in Iran or Southeast Asia. In fact, they almost certainly lived in South Asia for thousands of years before her.
The study also offers new insights into how farming began in South Asia. “Our findings do not prove a separate invention of farming in South Asia,” says geneticist David Reich from the US-based Harvard Medical School. Reich, who led a team of researchers from India, Germany and the US to sequence the genome, says their data is consistent with the theory that Western Asian agricultural technology or ideas moved into South Asia through adoption or ideas from neighbours.
Ancient human remains found in various sites of Indus Valley hardly yield intact DNA. The hot and humid conditions in these regions destroy any trace of DNA. To overcome this, Reich and post-doctoral scientist Vagheesh Narasimhan at Harvard, teamed up with Vasant Shinde from the Deccan College Post-Graduate and Research Institute in Pune and Niraj Rai from the Birbal Sahni Institute of Palaeosciences in Lucknow. They painstakingly screened 61 skeletal samples excavated from graves in Rakhigarhi and were eventually able to detect a very small amount of DNA in a single sample from a woman’s remains.
After more than hundred attempts, they were able to sequence the DNA. Comparing this DNA with those of 11 individuals from two sites in Turkmenistan and Iran, the researchers prepared a genetic profile of the Rakhigarhi woman.
The profile, they say, has signs of Iranian-related ancestry but no evidence of pastoralists who lived in the grasslands of Asia and Europe. “We say ‘Iranian-related’ because we don’t know where they lived,” Reich says. They could have lived in the Iranian plateau, but the team’s data point to them having lived in South Asia for many thousands of years before the Indus Valley Civilisation, he adds.
“This is a significant technical feat,” says Partha Pratim Majumder, an expert on human evolution and migration at the National Institute of Biomedical Genomics in West Bengal, India. “Sequencing genomes from South Asia has been a major challenge,” he adds.
This single sequence, he says, has provided a deep, though likely incomplete, insight into the Indus Valley lineage.
The analysis by Reich and colleagues also shows that the Iranian-related lineage present in the Indus Valley people split from the natives of Zagros Mountains in Iran before 8000 BCE. This is before crop farming began there around 7000–6000 BCE.
This suggests that the descendants of the world’s first farmers who lived in the Fertile Crescent have had no roles in introducing farming to South Asia.
Ancient DNA studies have shown that the hunter-gatherers in western Anatolia, a region in modern-day Turkey, adopted agriculture from their neighbours in the east. They then spread agriculture as they moved into Europe.
“Something similar might have happened in the vicinity of South Asia, where a hunter-gatherer population could have copied farming innovations from their eastern neighbors, and then spread them further through movement of people,” Reich points out.
There is little doubt that farming arose multiple times in different regions of the world, including in India, contends Majumder. Fossilized pollen grains, he says, point to multiple plant domestication and farming events in India. Thus, South Asia, in particular India, has always been a prominent seat of ancient farming, he adds.
The Indus Valley people built a complex and cosmopolitan culture, and variations are hard to decipher by decoding the genome of an individual, the researchers note. However, the insights that emerge from an individual genome demonstrate the promise of ancient DNA studies in South Asia, they add.
1. Shinde, V. et al. An ancient Harappan genome lacks ancestry from Steppe pastoralists or Iranian farmers. Cell. 179, 1-7 (2019) doi: 10.1016/j.cell.2019.08.048
doi: https://doi.org/10.1038/nindia.2019.121
Reprints and permissions
Assistant Professor in Integrated Photonics
We offer you the chance to design a unique and autonomous research program, networking with specialists, students and entrepreneurs.
Gothenburg (Stad), Västra Götaland (SE)
Chalmers University of Technology
Postdoctoral Fellow (Aging, Metabolic stress, Lipid sensing, Brain Injury)
Seeking a Postdoctoral Fellow to apply advanced knowledge & skills to generate insights into aging, metabolic stress, lipid sensing, & brain Injury.
Dallas, Texas (US)
UT Southwestern Medical Center - Douglas Laboratory
High-Level Talents at the First Affiliated Hospital of Nanchang University
For clinical medicine and basic medicine; basic research of emerging inter-disciplines and medical big data.
Nanchang, Jiangxi, China
The First Affiliated Hospital of Nanchang University
POSTDOCTORAL Fellow -- DEPARTMENT OF Surgery – BIDMC, Harvard Medical School
The Division of Urologic Surgery in the Department of Surgery at Beth Israel Deaconess Medical Center and Harvard Medical School invites applicatio...
Boston, Massachusetts (US)
Director of Research
Applications are invited for the post of Director of Research at Cancer Institute (WIA), Chennai, India.
Chennai, Tamil Nadu (IN)
Cancer Institute (W.I.A)
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Subscribe Now! Get features like

- Latest News
- Entertainment
- Real Estate
- Election Schedule 2024
- Win iPhone 15
- IPL 2024 Schedule
- IPL Points Table
- IPL Purple Cap
- IPL Orange Cap
- Bihar Board Results
- The Interview
- Web Stories
- Virat Kohli
- Mumbai News
- Bengaluru News
- Daily Digest

Harappan city: DNA samples of 2 skeletons now sent for analysis
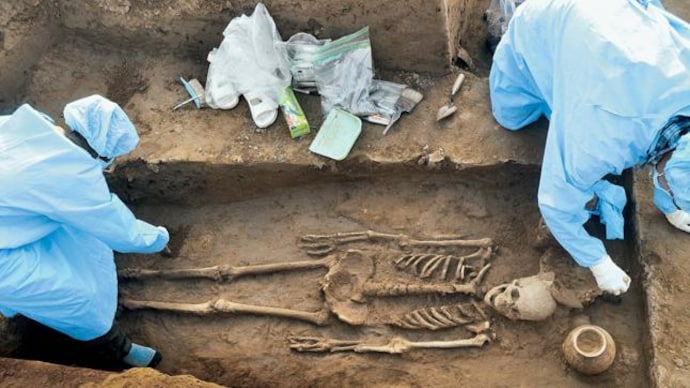
Dna samples collected from two human skeletons unearthed at a necropolis of a harappan-era city site in haryana have been sent for scientific examination, the outcome of which might tell about the ancestry and food habits of people who lived in rakhigarhi region thousands of years ago.
DNA samples collected from two human skeletons unearthed at a necropolis of a Harappan-era city site in Haryana have been sent for scientific examination, the outcome of which might tell about the ancestry and food habits of people who lived in Rakhigarhi region thousands of years ago.

The skeletons of two women were found a couple of months ago at mound number 7 (named RGR 7 by the Archaeological Survey of India), believed to be nearly 5,000 years old. Pots and other artefacts were also found buried next to them in a pit, part of the funerary rituals back in the Harappan Civilisation era, ASI officials said.
“Seven mounds (RGR 1- RGR 7) scattered around two villages (Rakhi Khas and Rakhi Shahpur) in Hisar district are part of the Rakhigarhi archaeological site. RGR 7 is a cemetery site of the Harappan period when this was a well-organised city. The two skeletons were unearthed about two months ago by our team. And, DNA samples were collected by experts about two weeks ago,” Joint Director General, ASI, S K Manjul told PTI.
At present RGR 1, RGR 3 and RGR 7 have been taken up for investigation.
Manjul, who is leading the excavation team at Rakhigarhi site, about 150 km north-west of Delhi, since it commenced on February 24, 2022, said the DNA analysis will help answer a lot of questions, anthropological or otherwise.
The samples will be first examined by Birbal Sahni Institute of Paleosciences, Lucknow for preliminary investigation and scientific comparison, before being sent further for forensic analysis from anthropological perspective, he said.
“The outcome of the DNA analysis will help tell about the ancestry of the people who lived at this ancient city, whether they were native or had migrated from elsewhere to settle. Besides, samples taken from the teeth area would tell about their food habits, what kind of food they consumed and other anthropological patterns related to that human settlement which must have been one of the largest, dating from the Harappan Civilisation period,” said Manjul, who had also led the excavation at Sanauli in Uttar Pradesh in 2018 where pre-Iron Age artefacts were unearthed.
For collection of DNA samples, experts had done it while wearing special uniform so as to not contaminate the samples. And, samples were taken from teeth region and petrous part of the temporal bone, located at the base of the skull in the ear region.
Rakhigarhi site is one of the “five iconic sites” declared by the central government as per the Union budget 2020-21.
The two skeletons were found lying in supine position with head pointing in the north direction. They both were buried with plethora of pottery and adorned jewellery like jasper and agate beads and shell bangles. A symbolic miniature copper mirror was found buried along with one of the skeletons, officials said.
Animal bones were also found at the site, they said.
First attempts to archaeologically explore the Rakhigarhi site is said to have been done in late 1960s.
Join Hindustan Times
Create free account and unlock exciting features like.

- Terms of use
- Privacy policy
- Weather Today
- HT Newsletters
- Subscription
- Print Ad Rates
- Code of Ethics
- Elections 2024
- India vs England
- T20 World Cup 2024 Schedule
- IPL Live Score
- IPL 2024 Auctions
- T20 World Cup 2024
- Cricket Players
- ICC Rankings
- Cricket Schedule
- Other Cities
- Income Tax Calculator
- Budget 2024
- Petrol Prices
- Diesel Prices
- Silver Rate
- Relationships
- Art and Culture
- Telugu Cinema
- Tamil Cinema
- Exam Results
- Competitive Exams
- Board Exams
- BBA Colleges
- Engineering Colleges
- Medical Colleges
- BCA Colleges
- Medical Exams
- Engineering Exams
- Horoscope 2024
- Festive Calendar 2024
- Compatibility Calculator
- The Economist Articles
- Explainer Video
- On The Record
- Vikram Chandra Daily Wrap
- PBKS vs DC Live Score
- KKR vs SRH Live Score
- EPL 2023-24
- ISL 2023-24
- Asian Games 2023
- Public Health
- Economic Policy
- International Affairs
- Climate Change
- Gender Equality
- future tech
- Daily Sudoku
- Daily Crossword
- Daily Word Jumble
- HT Friday Finance
- Explore Hindustan Times
- Privacy Policy
- Terms of Use
- Subscription - Terms of Use
- Law & Policy
- Society & Culture
- Business & Economy
- Science & Technology
- Daan & Sewa
- Hindu Human Rights
- Write For Us
HinduPost is the voice of Hindus. Support us. Protect Dharma

Will you help us hit our goal?
We’re aiming to add 2,500 contributions in the next 30 days, to help keep HinduPost free.
Rakhigarhi DNA Analysis: busting the Aryan Invasion myth
“A lie can travel half way around the world while the truth is putting on its shoes” – Mark Twain.
For centuries, a lie known as the ‘ARYAN INVASION THEORY (AIT)’ was first propagated by the British colonizers and later passed on by various vested interests, based entirely on false science, assumptions & convoluted logic.
We owe this web of lies to the questionable linguist Max Mueller and archaeologist Mortimer Wheeler who conveniently ignored available artefacts, mis-interpreted Sanskrit Vedic texts and tried to force-fit events as per a Biblical timeline. Following their lead, many indigenous and foreign archaeologists, historians and writers continued propagating this theory, some out of ignorance and others simply because it suited their agenda.
But as they say, ‘you can run with a lie but you can’t hide from the truth. Eventually truth will catch up’. And that is what has happened! As scientific advancements are made, a lot of evidence has stumbled out which prove that AIT was a misplaced concept that had no basis in reality.
As more and more sites of what is actually Saraswati-Sindhu Civilization (SSC) are being dug up, overwhelming archaeological, linguistic and other evidence is coming out in the public domain, thus rendering the AIT irrelevant, false and outdated.
Now, internationally renowned archaeologist Prof. Vasant Shinde of Deccan College, Pune has discovered 5000-year-old DNA evidence, from the largest SSC site of Rakhigarhi in Haryana, that conclusively proves that present day Bharatiyas are direct descendants of the SSC people and that so called Aryans neither invaded nor migrated to Bharat. DNA and other evidence from Rakhigarhi indicates that SSC was indeed the Vedic civilization that continues to be practised even today.

Rakhigarhi in Haryana is arguably the most important SSC site discovered till now. Spread over 550 hectares, it is double the size of Mohenjo Daro. Ongoing excavations at this over 5000-year-old site have provided evidences of the place being a major manufacturing and trade hub, and study of skeletal remains showed no sign of violent conflict and a manner of burial quite similar to the early Vedic period.
In 2016 interview, Prof. Vasant Shinde had said “Scientists have, for the first time ever, succeeded in extracting DNA from the skeletons of the Indus Valley Civilisation. More skeletons have been found during the ongoing excavation season from mound 2 for further analysis. Three different institutes of world repute are conducting the DNA analysis for a foolproof study, so there is no scope of any contradiction”.
A team led by geneticist David Reich at Harvard University succeeded in obtaining DNA signature from a few skeletal remains. They developed a new scientific technique to extract and study DNA from the petrous bone of the inner ear of a 5000-year-old skeleton of a woman. The results of the aforementioned DNA have brought to light some very interesting findings and have established firmly that AIT was nothing but a myth. Here are some of these findings:
1) No DNA strands either from Iranian or Steppe pastoralists in the Rakhigarhi sample shows that these people were indigenous and they independently developed farming practices in Rakhigarhi.
2) DNA of the Rakhigarhi sample matches with those of modern Bharatiyas.
3) Continuity of culture is visible from Rakhigarhi to modern times which is seen in both the genetic and archaeological data. Urbanization was introduced by these indigenous people.
Other archaeological sites
Several archaeological sites have thrown up ample evidence that points towards indigenous origins of Bharatiya civilization and developments starting from as early as 7000 BCE. These sites such as Mehrgarh, Girawad, Mitathal, Farmana, Kunal etc are strewn across the Saraswati-Sindhu river basin on both sides of the border, and now sites are also being found in Gangetic plains and places like Odisha.
Fire altars have been found at 5700-year-old Lothal SSC site in Gujarat that relate to ancient Vedic Yagna practices. So archaeological & genetic evidence are both now saying that no ‘Aryans’ invaded or migrated into ancient Bharat and that indigenous people, ancestors of present day Bharatiyas, have been inhabiting this land since times immemorial.
Why was the Aryan Invasion Theory created?
For a long time now Aryan Invasion Theory (AIT) or its variant Aryan Migration Theory (AMT), has been passed off as the truth. It started with the likes of colonial and western archaeologists, linguists and ‘historians’ such as Mortimer Wheeler, Max Mueller and F. Bopp among others. Nehru in his book ‘Discovery of India’ and Lokmanya Tilak also talked of Aryans having come from outside. Dr. Ambedkar, however, strongly refuted this theory in his book ‘Who were Shudras?’.
Initially, it was said that Aryans invaded Bharat and pushed the indigenous people towards the South. When no traces of war or evidence supporting this view was found, which was simply based on finding one pile of skeletons, propagators of this view began to say the Aryans migrated in waves into Bharat and imported their Vedic knowledge and lifestyle which were alien to original inhabitants of Bharat.
AIT gave a historical precedent to justify the role and status of the British Raj, who could argue that they were transforming Bharat for the better in the same way that the Aryans had done thousands of years earlier.
The AIT premise was also used as a perfect tool by the British to divide the Hindu society and the state of Bharat. The North Indian “Aryans” were then pitted against the South Indian “Dravidians,” along with high-caste against low-caste, mainstream Hindus against tribals, Vedic orthodoxy against the indigenous orthodox sects, and later to neutralize Hindu criticism of the forced Islamic occupation of Bharat, since “Hindus themselves entered Bharat in the same way as Muslims did.” Even today, the theory has still been used as the basis for the growth of secularist, Marxist and other #BreakingIndia forces like Dravidianists. Even some feudalistic Hindus have bought into the theory to prove their ‘superiority’ over other Hindus.
These DNA results not just reject Aryan invasion/migration theory, but also prove that hunter-gatherers who roamed Bharat were the ones who actually founded the early Sindhu-Saraswati Civilization (SSC) somewhere around 7000 BCE or more. With time, a material culture developed giving rise to an urban civilization, but the genetic makeup remained the same.
It has also been established that Vedic culture was indeed developed by indigenous people in the Sindhu Saraswati basin and that continues to live even today, making Hindu Vedic culture the oldest surviving continuous culture.
This is but the beginning of a story that would unravel more mysteries as more studies are conducted and evidence unearthed. While we are unearthing new evidence, it is perhaps time to safely bury the Aryan Invasion/Migration myth that has been propagated for far too long to serve the agenda of a few.
https://www.cell.com/cell/fulltext/S0092-8674(19)30967-5
https://www.sciencemag.org/news/2019/09/genome-nearly-5000-year-old-woman-links-modern-indians-ancient-civilization
https://science.sciencemag.org/content/365/6457/eaat7487
https://www.tribuneindia.com/news/haryana/rakhigarhi-more-important-than-mohenjo-daro-data/231004.html
https://hindupost.in/6000-year-old-rakhigarhi-burials-were-done-in-rig-vedic-fashion-dna-study-finds-no-central-asian-trace/
https://www.stephen-knapp.com/aryan_invasion_theory_the_final_nail_in_its_coffin.htm
https://timesofindia.indiatimes.com/city/kolkata/Varanasi-is-as-old-as-Indus-valley-civilization-finds-IIT-KGP-study/articleshow/51146196.cms
http://www.asianage.com/india/all-india/260517/artefacts-of-4000-year-old-civilisation-found-in-odisha.html
Did you find this article useful? We’re a non-profit. Make a donation and help pay for our journalism.
Subscribe to our channels on Telegram & YouTube . Follow us on Twitter and Facebook
Related Articles
Bharatavarsha must consciously reject the western model of ideological universalism and accelerate decolonisation, decolonisation is a national duty and a hindu civilisational impertive, liberating justice from colonial legacy: decoding the nyaya, suraksha & sakshya bills.
- Archaeology
- Aryan Invasion Theory
- Colonial Legacy
- Decolonisation
- Reclaim History
- Sindhu-Saraswati Valley Civilisation
- Western Indology
LEAVE A REPLY Cancel reply
Save my name, email, and website in this browser for the next time I comment.
Yes, add me to your mailing list
Latest Articles
Secret history of caste, fascinating collection from the church, gujarat: vadodara based famous ‘huseni samosa’ used to actually sell beef-stuffed samosas; yusuf, naeem, and four others arrested, one of the most engaging but dishonest film is jana gana mana, from commiewood, supreme court defends youtuber’s right to criticise tamil nadu cm stalin, restores bail in defamation case, ‘received threat letter from let on nrc’, claims union minister shantanu thakur.

Sign up to receive HinduPost content in your inbox
We don’t spam! Read our privacy policy for more info.
Check your inbox or spam folder to confirm your subscription.

HinduPost is a news, commentary and opinion digital media outlet that provides the correct perspective on issues concerning Hindu society.
Popular Tags
COPYRIGHT © HINDUPOST.IN
- Privacy Policy
- Terms & Conditions

Thanks for Visiting Hindupost
- Prehistoric
- From History
- Cultural Icons
- Women In history
- Freedom fighters
- Quirky History
- Geology and Natural History
- Religious Places
- Heritage Sites
- Archaeological Sites
- Handcrafted For You
- Food History
- Arts of India
- Weaves of India
- Folklore and Mythology
- State of our Monuments
- Conservation

New Findings at Rakhigarhi
- AUTHOR Disha Ahluwalia
- PUBLISHED 21 May 2022
Rakhigarhi in Haryana is one of the most important Harappan sites in India. A recent phase of excavations had made national headlines, thanks to fascinating discoveries such as a lapidary (bead-making centre), drains and a burial ground. This has led to an increased understanding of Rakhigarhi as a site and of Harappan settlements in India. Disha Ahluwalia, an archaeologist and excavation supervisor at Rakhigarhi excavation, shares some very interesting insights into these recent discoveries
On 5th January 1921, Rai Bahadur Daya Ram Sahni began excavating at Harappa, on the banks of the Ravi River. Little did he know that this excavation would push back the antiquity of the Indian subcontinent to the 3rd millennium BCE, making the newly found civilization a contemporary of the Mesopotamian and Egyptian civilizations.
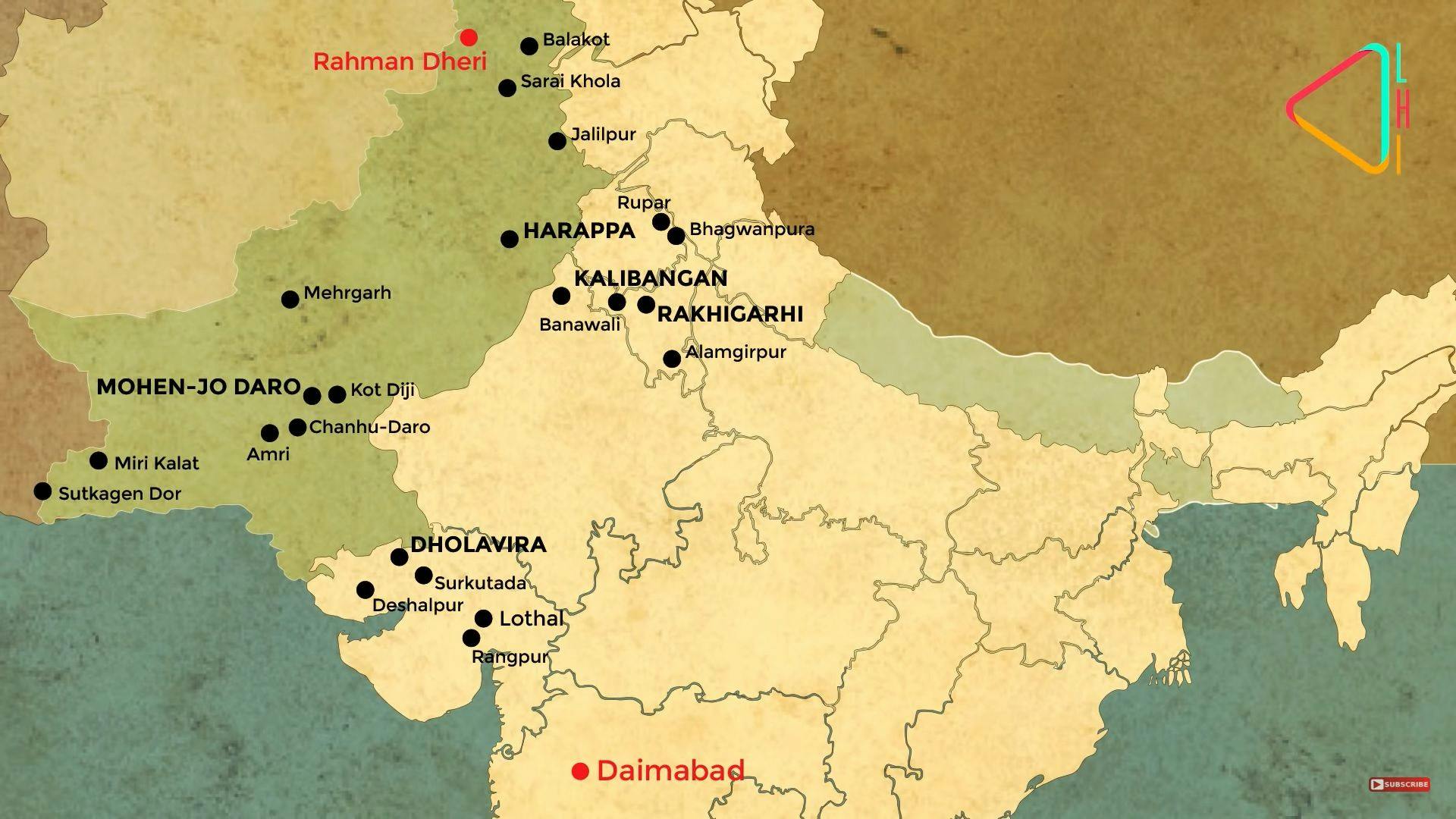
The Harappan Civilization, as it is now known (named after the first archaeological site that marked the discovery of this Civilization) is the largest civilization of the ancient world. Spread from Sutkagan Dor in the west to Alamgirpur in the east, it is not restricted by one river valley or current political boundaries. A century has gone into excavating, understanding and assessing the remains from the sites that dot this landscape. We now know that the Harappans lived in a civilization that was dynamic, diverse yet unified. It comprises a mix of rural, semi-rural, factory/industrial and urban centres.
Of these, five major cities, the metros of Mohenjo-daro, Harappa, Ganweriwal, Rakhigarhi and Dholavira , stand out due to their sheer size and magnitude. These big cities are known for the straified settlements divided into the citadel, middle town and lower town; systematic town planning; drainage systems; rainwater harvesting; etc.
Each has its own unique features yet unified by standardisation of brick size, weights and measures, to name a few. More importantly, these major cities are located strategically on the trade route of the Harappans, covering the different geographical zones within the realm of the civilization.
Rakhigarhi – An Iconic Site

Mohenjo-daro and Harappa on the Indus River Valley and Dholavira in Kutch, Gujarat, have been extensively excavated. Rakhigarhi, on the other hand, being the largest site in the eastern (Indian) domain of the civilization and on the banks of the Chautang of the Saraswati river system, remains largely unexcavated. Comprising seven mounds (RGR 1 to 7), it is spread across the present-day villages of Rakhi Khas and Rakhi Shahpur and adjoining fields in Hissar district of Haryana, merely 160 km west of New Delhi.
The archaeological wealth of the site was first reported by Dr Suraj Bhan in 1969, but the site was subjected to systematic excavation in 1998 for the first time. He collected Mature Harappan (c. 2600-1900 BCE) artifacts from the surface, which suggested that the site is at least 5,000 years old.
But the first systematic excavation was carried out by the Institute of Archaeology, ASI, under the directorship of Dr Amrinder Nath, who carried out three field seasons (1998-2001) on all seven mounds. A large number of important antiquities including Harappan seals were excavated along with structural remains.

This excavation gave a broad chronological context to the site, right from c. 4000 BCE to 1900 BCE. In other words, it suggested that Rakhigarhi as a city in the Mature Harappan phase evolved from a largely agrarian, Early Harappan phase. Excavations were also carried out in the burial ground (RGR 7) that unearthed many skeletal remains of the Harappan period, an example of which you can see in the National Museum, New Delhi.
The importance of this excavation goes beyond just retrieving archaeological data as it laid a strong foundation for future endeavours. After a hiatus of over a decade, the site was re-excavated by Deccan College, Pune, headed by Prof Vasant Shinde in collaboration with the Haryana State Archaeology Department, from 2013-16. The famous DNA study was conducted during this time, and it grabbed headlines and made the world look at Rakhigarhi from a fresh perspective.
Excavations at Rakhigarhi
Before the pandemic paralyzed the globe, the Government of India had declared around Rs 3,000 crore for the development five major archaeological sites – Dholavira, Adhichanallur, Hastinapur, Shiv Sagar and Rakhigarhi. The aim of this project is simple – to promote the relics of the past, to educate people about the glorious past of the country, to provide amenities to visitors, and to conserve the sites for public viewing. The project aimed at promoting the relics of the past. Taking this lead, the Archaeological Survey of India started excavation at Rakhigarhi on 24th February 2022 under the directorship of Dr Sanjay Manjul, on the lines of developing this major Harappan city under the government project and most importantly to understand the settlement of Rakhigarhi, holistically by identifying the individuality and interrelationship of the seven mounds.

Three of the seven mounds, RGR 1, 3 and 7, were taken up for investigation by Dr Manjul and his team.
The Lapidary Cent
Upon reaching Rakhigarhi in mid-February 2022, I set foot on this 6m high mound, at the corner of which our camp was set. As I put down my bags in my tent, I looked out at this site which we were about to break open with our trowels. We started with digging trenches on the eastern half of the mound. Earlier, the western half of RGR 1 was excavated by Dr Nath and his team in 1998-01. They uncovered a large quantity of debitage/waste of semi-precious stones such as agate, carnelian and jasper, which was used in the manufacture of objects like beads. They also reported a furnace and hundreds of semi-finished beads. Taking clues from that data, we wished to further the information on the nature of this site aided with scientific means.

Only a few centimeters below the ground, structural remains began to appear and soon we found ourselves digging a Harappan street about 2.6 m wide and running for 18 m in an east-west orientation. The street was being used for a long time and was raised as the level of habitation was rising. The street is between two mud-brick structures. These lanes were found intersecting at 90 degrees. Interestingly, at the intersections, soakage jars were placed, an attempt by the Harappans to keep their city clean.
It was thrilling to uncover the kind of evidence we grew up reading in textbooks. The street with a multiple coursed brick wall that was 18m in length and evidence of workshops-cum-habitation on either side evidently suggested that Rakhigarhi was a well-planned city with high standards of Harappan town planning.

Besides this, semi-finished and finished beads, a large quantity of debitage/waste, stones, raw material, hearths etc strongly suggest that at RGR 1 Lapidary was the main activity and it was a settlement of craftsmen.
Street with Drain

With a blue marbled Mazar on top of the mound, RGR 3 is sandwiched between two of Rakhigarhi’s largest mounds – RGR 2 on the west and RGR 4 on the east and south-west of RGR 1. This mound is approximately 11 m high and remained unexcavated till Dr Manjul decided to investigate the site to tap into the ‘grey areas of research’.
Evidence of a baked brick wall about 15 m in length, enclosing a residential complex and a brick-lined drain on the one side is yet another example of sophisticated town-planning that the Harappans are known for. The kind of antiquities and pottery from this mound differentiate its nature from RGR 1 but at this point in research, it’s hard to come to any conclusion.
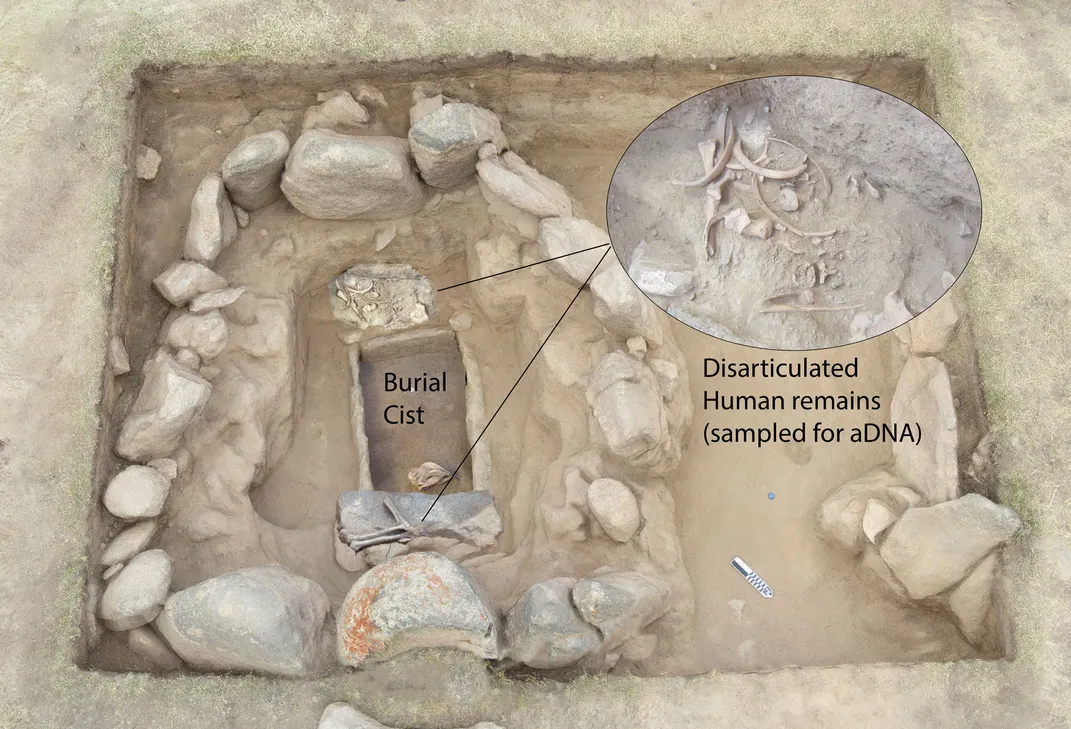
The Burial Ground

The DNA sample retrieved from Rakhigarhi gave an impetus to a long-drawn debate and controversy. This sample was extracted from one of over 60 burials excavated at RGR 7 located about 500 m north of the lapidary centre, (RGR 1) in the agricultural field. Buried in privately owned agriculture fields, the remains of this ‘cemetery’ of the people of Rakhigarhi yielded a large number of skeletal remains unearthed in 1998-01 and then in 2013-16.
However, despite the DNA results and multiple seasons of excavation, there was a need to link this ‘cemetery’ to the rest of the six mounds on the basis of material culture and also to see if multiple phases of burial activity was prevalent in the ancient past.
After a few weeks of digging, two burials of female individuals were unearthed. They were buried with a plethora of pottery and adorned jewelry like jasper and agate beads and shell bangles. In one burial, a symbolic miniature copper mirror was buried along with the skeleton.

Since these burials were found at the topmost level, an attempt was made to dig below in one portion of the trench. Instead of more skeletons, we uncovered habitation levels that were not known to us before. Multiple hearths at different working levels pointed to an earlier settlement at RGR 7. This raises many questions that will be answered in the coming field season.

From streets to drainage and a plethora of important antiquities including Harappan seals and a sealing with an elephant relief, the celebrated past of this archaeological site has once again been narrated. We now know that the city was well planned, even the areas where craftsmen of the city resided. What we now also know is that trade and rising demand for finished goods enabled the city to grow not only as a trading centre but also as a manufacturing centre.
However what we now most certainly know is that before Rakhigarhi evolved into a big metropolis, the Early Harappan settlement was larger than previously thought, in other words the findings especially of early Habitation level at RGR 7, are motivating us to look beyond the set narratives of the past. As eminent archaeologist D P Aggarwal once said, the ' fear of new discoveries should not come in our way of reconstruction of the past… new data will always force revision; that is the way of all research'. Our only hope is that the next season will bring more clarity to the findings and will bring a new dawn to the past that is buried beneath the ground at Rakhigarhi.
If you enjoyed this article, you will love LHI Circle - your Digital Gateway to the Best of India's history and heritage. You can enjoy our virtual tours to the must-see sites across India, meet leading historians and best-selling authors, and enjoy tours of the top museums across the world. Join LHI Circle here
Handcrafted Home Decor For You

Blue Sparkle Handmade Mud Art Wall Hanging

Handcrafted Tissue Box Cover Sea Green & Indigo Blue (Set of 2)

Scarlet Finely Embroidered Silk Cushion Cover

Sunflower Handmade Mud Art Wall Hanging

Related Stories
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- HHS Author Manuscripts

An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers
Vasant shinde.
1 Department of Archaeology, Deccan College Post-Graduate and Research Institute, Pune 411006, India
Vagheesh Narasimhan
2 Department of Genetics, Harvard Medical School, Boston, MA 02115, USA
Nadin Rohland
Swapan mallick.
3 Howard Hughes Medical Institute, Harvard Medical School, Boston, MA 02115, USA
4 Broad Institute of Harvard and MIT, Cambridge, MA 02142, USA
Matthew Mah
Mark lipson, nathan nakatsuka, nicole adamski, nasreen broomandkoshbacht, matthew ferry, ann marie lawson, megan michel, jonas oppenheimer, kristin stewardson, nilesh jadhav, yong jun kim, malvika chaterjee, avradeep munshi, amrithavalli panyam, pranjali waghmare, yogesh yadav, himani patel.
5 Birbal Sahni Institute of Palaeosciences, Lucknow 226007, India
Amit Kaushik
6 Amity Institute of Biotechnology, Amity University, Sector125, Noida 201303, India
Kumarasamy Thangaraj
7 CSIR-Centre for Cellular and Molecular Biology, Hyderabad 500 007, India
Matthias Meyer
8 Max Planck Institute for Evolutionary Anthropology, Leipzig 04103, Germany
Nick Patterson
9 Department of Human Evolutionary Biology, Harvard University, Cambridge MA, 02138, USA
David Reich
10 Lead Contact
Author contributions
Associated Data
All newly reported sequencing data are available from the European Nucleotide Archive, accession number PRJEB34154.
We report an ancient genome from the Indus Valley Civilization (IVC). The individual we sequenced fits as a mixture of people related to ancient Iranians (the largest component) and Southeast Asian hunter-gatherers, a unique profile that matches ancient DNA from 11 genetic outliers from sites in Iran and Turkmenistan in cultural communication with the IVC. These individuals had little if any Steppe pastoralist-derived ancestry, showing it was not ubiquitous in northwest South Asia during the IVC as it is today. The Iranian-related ancestry in the IVC derives from a lineage leading to early Iranian farmers, herders and hunter-gatherers before their ancestors separated, contradicting the hypothesis that the shared ancestry between early Iranians and South Asians reflects a large-scale spread of western Iranian farmers east. Instead, sampled ancient genomes from the Iranian plateau and IVC descend from different groups of hunter-gatherers who began farming without being connected by movement of people.
Skeletal DNA from a member of the ancient Indus Valley Civilization shows ancestry from ancient Iranians before their adoption of farming and from Southeast Asian hunter-gatherers, while completely lacking Steppe pastoralist ancestry.
Graphical Abstract

Introduction
The mature Indus Valley Civilization (IVC), also known as the Harappan Civilization, was spread over northwestern South Asia from 2600–1900 BCE and was one of the first large-scale urban societies of the ancient world, characterized by systematic town planning, elaborate drainage systems, granaries, and standardization of weights and measures. The inhabitants of the IVC were cosmopolitan, with multiple cultural groups living together in large regional urban centres like Harappa (Punjab), Mohenjo-daro (Sindh), Rakhigarhi (Haryana), Dholavira (Kutch/Gujarat) and Ganweriwala (Cholistan) ( Figure 1A ) ( Mughal, 1990 ; Possehl, 1982 ; Possehl, 1990 ; Shaffer and Lichtenstein, 1989 ; Thapar, 1979 ). Rakhigarhi, the largest known IVC site, was spread over 550 hectares ( Figure 1B , ,C), C ), and seven dates from charcoal at depths of 9–23 meters have point estimates of 2800–2300 BCE, which largely fall within the mature phase of the IVC ( Shinde et al., 2018 ; Vahia et al., 2016 ). As part of the archaeological effort we attempted to generate ancient DNA data for a subset of the excavated burials.

( A ) We label the geographic location of the archaeological site of Rakhigarhi (blue) and other significant Harappan sites (red) to define the geographic range of the IVC. We label in black sites in the north and west with which IVC people were in cultural contact, and specifically highlight in yellow the sites of Gonur and Shahr-i-Sokhta, which are the source of the 11 outlier individuals who genetically form a cline of which the Rakhigarhi individual is a part. ( B ) Photograph of the I6113 burial (skeletal code RGR7.3, BR-01, HS-02) and associated typical IVC grave goods and illustrating typical North-South orientation of IVC burials. High resolution images of IVC-style ceramics associated with the grave are shown in Figure S1 .
In dedicated clean rooms, we obtained powder from 61 skeletal samples from the Rakhigarhi cemetery, which lies ~1 km west of the ancient town ( Table S1 ). We extracted DNA ( Dabney et al., 2013 ; Korlević et al., 2015 ) and converted the extracts into libraries ( Rohland et al., 2015 ), some of which we treated with uracil-DNA glycosylase (UDG) to greatly reduce the error rates associated with the characteristic cystosine-to-uracil lesions of ancient DNA ( Rohland et al., 2015 ). For a subset of libraries that passed basic laboratory quality controls showing they contained DNA, we enriched for sequences overlapping both the mitochondrial genome and ~3000 targeted nuclear positions ( Mathieson et al., 2015 ), and sequenced the enriched libraries either on Illumina NextSeq500 instrument using paired 2×76 base pair (bp) reads, or on Illumina HiSeq X10 instruments using paired 2×150 bp reads. After trimming adapters and merging sequences overlapping by at least 15 bp (allowing up to one mismatch), we mapped to both the mitochondrial genome rsrs ( Behar et al., 2012 ) and the human genome reference hg19 ( Li and Durbin, 2010 ) ( Table S1 ). After inspecting the screening results we enriched a subset of libraries for ~1.24 million single nucleotide polymorphisms (SNPs), and sequenced the enriched libraries and processed the data computationally ( Fu et al., 2015 ; Haak et al., 2015 ; Mathieson et al., 2015 ). For the most promising sample, which had the genetic identification code I6113 and the archaeological skeletal code RGR7.3, BR-01, HS-02, we created, enriched, and sequenced a total of 108 libraries from 5 extractions to maximize the amount of data ( Meyer et al., 2012 ; Rohland et al., 2015 ) (the initial library was UDG-treated, while all 107 subsequent libraries were not UDG-treated). After removing 40 libraries (from one extraction) that had significantly lower coverage as well as significantly lower damage rates compares to the other libraries, and merging data from the remaining 68 libraries, we had 208,111 SNPs covered at least once. Almost all of these 68 libraries showed cytosine-to-thymine mismatch rates to the human reference genome in the final nucleotide greater than 10%, consistent with the presence of authentic ancient DNA. However, when we stratified the pooled data by sequence length we found lower damage rates particularly for sequences of length >50 bp ( Star Methods ). Based on this evidence of length-dependent contamination, we restricted the data to molecules that showed cytosine-to-thymine mismatches characteristic of ancient DNA. This resulted in data at 31,760 SNPs. The ratio of sequences mapping to the Y chromosome to sequences mapping to both the Y and X chromosomes was in the range expected for a female, consistent with the morphology of the skeleton. After building a mitochondrial DNA consensus sequence, we determined that its haplogroup was U2b2, which is absent in data from about 400 ancient Central Asians and today is nearly exclusive to South Asia ( Narasimhan et al., 2019 ).
In PCA ( Figure 2A ), I6113 projects onto a previously defined genetic gradient represented in 11 individuals from two sites in Central Asia in cultural contact with the IVC (3 from Gonur in present-day Turkmenistan and 8 from Shahr-i-Sokhta in far eastern Iran); these individuals were previously identified via a formal statistical procedure as significant outliers relative to the majority of samples at these two sites (they represent only 25% of the total) and were called the Indus Periphery Cline ( Narasimhan et al., 2019 ). Despite having only modest SNP coverage, the error bars for the positioning of I6113 in the PCA are sufficiently small to show that this individual is not only significantly different in ancestry from the primary ancient populations of Bronze Age Gonur and Shahr-i-Sokhta, but also does not fall within the variation of the present-day South Asians. We obtained qualitatively consistent results when analyzing the data using ADMIXTURE ( Alexander et al., 2009 ), with I6113 again similar to the 11 outlier individuals in harboring a mixture of ancestry related to ancient Iranians and tribal southern Indians. None of these individuals had evidence of “Anatolian farmer-related” ancestry, a term we use to refer to the lineage found in ancient genomes from 7 th millennium BCE farmers from Anatolia ( Mathieson et al., 2015 ). This Anatolian farmer-related ancestry was absent in all sampled ancient genomes from Iranian herders or hunter-gatherers dating from the 12 th through the 8 th millennia BCE, which instead carried a very different ancestry profile also present in mixed form in South Asia that we call “Iranian-related” ( Broushaki et al., 2016 ; Lazaridis et al., 2016 ).

( A ) PCA of ancient DNA from South and Central Asia projected onto a basis of whole genome sequencing data from present-day Eurasians. I6113 and {"type":"entrez-protein","attrs":{"text":"I11042","term_id":"592443","term_text":"I11042"}} I11042 (a non-outlier individual from the site of Gonur of similar data quality), are shown along with black error bars indicating 1 standard error as estimated using a chromosomal block jackknife. I6113’s position in the PCA is inconsistent with that of present-day South Asians and with Iranian groups, but is consistent with a set of 11 outliers who represent 25% of analyzed individuals at the sites of Gonur and Shahr-i-Sokhta and who with I6113 for the IVC Cline . ( B ) ADMIXTURE analysis of individuals from South and Central Asia shown with components in Green, Teal, Orange, Blue and Red maximized in Iranian farmers, Anatolian farmers, Eastern European Hunter-Gatherers, Western European Hunter-Gatherers, and Andamanese Hunter-Gatherers, respectively. ( C ) Estimated proportions of three ancestry profiles in ancient and present-day individuals. The three components are maximized in Middle to Late Bronze Age Steppe Pastoralists ( Central_Steppe_MLBA ), the reconstructed hunter-gatherer population of South Asia (represented by Andamanese Hunter-Gatherers (AHG) as proxy for with greatest relatedness to southeast Asian hunter-gatherers), and Indus_Periphery_West , an individual on the IVC Cline represented with the lowest proportion of AHG -related ancestry. Individuals that fit a two-way model of mixture between these three sources are shown on the triangle edges, whereas individuals that could only be fit with a three way model are shown in the interior. I6113 is shown on the IVC Cline as a red dot; the other IVC cline individuals are shown as orange dots; later individuals who formed as mixtures between people on the IVC Cline and people with Steppe ancestry are shown as green dots; and diverse modern South Asian groups who formed as a mixture of two later mixed groups are shown as blue dots
We used qpAdm to test highly divergent populations that have been shown to be effective for modeling diverse West and South Eurasian groups as potential sources for I6113 ( Narasimhan et al., 2019 ). If one of these population fits, it does not mean it is the true source; instead, it means that it and the true source population are consistent with descending without mixture from the same homogeneous ancestral population that potentially lived thousands of years before. The only fitting two-way models were mixtures of a group related to herders from the western Zagros mountains of Iran, and also to either Andamanese Hunter-Gatherers (73 ± 6% Iranian-related ancestry; p=0.103 for overall model fit) or East Siberian Hunter-Gatherers (63 ± 6% Iranian-related ancestry; p=0.24 (the fact that the latter two populations both fit reflects that they have the same phylogenetic relationship to the non-West Eurasian-related component of I6113 likely due to shared ancestry deeply in time). This is the same class of models previously shown to fit the 11 outliers that form the Indus Periphery Cline ( Narasimhan et al., 2019 ), and indeed, I6113 fits as a genetic clade with the pool of Indus Periphery Cline individuals in qpAdm (p=0.42). Multiple lines of evidence suggest that the genetic similarity of I6113 to the Indus Periphery Cline individuals is due to gene flow from South Asia rather than in the reverse direction. First, of the 44 individuals with good quality data we have from Gonur and Shahr-i-Sokhta, only 11 (25%) have this ancestry profile; it would be surprising to see this ancestry profile in the one individual we analyzed from Rakhigarhi if it was a migrant from regions where this ancestry profile was rare. Second, of the three individuals at Shahr-i-Sokhta that have material culture linkages to Baluchistan in South Asia, all are IVC Cline outliers, specifically pointing to movement out of South Asia ( Narasimhan et al., 2019 ). Third, both the IVC Cline individuals and the Rakhigarhi individual have admixture from people related to present-day South Asians (ancestry deeply related to Andamanese hunter-gatherers) that is absent in the non-outlier Shahr-i-Sokhta samples and is also absent in Copper Age Turkmenistan and Uzbekistan ( Narasimhan et al., 2019 ), implying gene flow from South Asia into Shahr-i-Sokhta and Gonur, whereas our modeling does not necessitate reverse gene flow. Based on these multiple lines of evidence it is reasonable to conclude that individual I6113’s ancestry profile was widespread among people of the IVC at sites like Rakhigarhi, and supports the conjecture ( Narasimhan et al., 2019 ), that the 11 outlier individuals in the Indus Periphery Cline are migrants from the IVC living in non-IVC towns. We rename the genetic gradient represented in the combined set of 12 individuals the “ IVC Cline ,” and then use higher coverage individuals from this cline in lieu of I6113 to carry out fine-scale modeling of this ancestry profile.
Modeling the individuals on the IVC Cline using the two-way models previously fit for diverse present-day South Asians ( Narasimhan et al., 2019 ), we find that as expected from the PCA it does not fit the two-way mixture that drives variation in modern South Asians as it is significantly depleted in Steppe pastoralist-related ancestry adjusting for its proportion of Iranian-related ancestry (p=0.018 from a two-sided Z-test). Modeling the IVC Cline using the simpler two-way admixture model without Steppe pastoralist-derived ancestry previously shown to fit the 11 outliers ( Narasimhan et al., 2019 ), I6113 falls on the more Iranian-related end of the gradient, revealing that Iranian-related ancestry extended to the eastern geographic extreme of the IVC, and was not restricted to individuals at its Iranian and Central Asian periphery. The estimated proportion of ancestry related to tribal groups in southern India in I6113 is smaller than in present-day groups, suggesting that since the time of the IVC there has been gene flow into the part of South Asia where Rakhigarhi lies from both the northwest (bringing more Steppe ancestry) and southeast (bringing more ancestry related to tribal groups in southern India). The genetic profile that we document in this individual, with large proportions of Iranian-related ancestry, but no evidence of Steppe pastoralist-related ancestry, is no longer found in modern populations of South Asia or Iran, providing further validation that the data we obtained from this individual reflects authentic ancient DNA.
To obtain insight into the origin of the Iranian-related ancestry in the IVC Cline , we co-modeled the highest-coverage individual from the IVC Cline (who also happens to have the highest proportion of Iranian-related ancestry) with other ancient individuals from across the Iranian plateau representing early hunter-gatherer and food producing groups: a ~10000 BCE individual from Belt Cave in the Alborsz Mountains, a pool of ~8000 BCE early goat herders from Ganj Dareh in the Zagros Mountains, a pool of ~6000 BCE farmers from Hajji Firuz in the Zagros Mountains, and a pool of ~4000 BCE farmers from Tepe Hissar in Central Iran. Using qpGraph ( Patterson et al., 2012 ), we tested all possible simple trees relating the Iranian-related ancestry component of these groups, accounting for known admixtures (Anatolian farmer-related admixture into Hajji Firuz and Tepe Hissar, and Andamanese Hunter-Gatherer-related admixture in the IVC Cline ), using an acceptance criterion for the model fitting that the maximum |Z|-scores between observed and expected f -statistics was <3, or that the Akaike Information Criterion (AIC) was within 4 of the best-fit ( Burnham and Anderson, 2004 ). The only consistently fitting models specified the IVC Cline Iranian-related ancestry lineage as splitting before the other Iranian-related lineages separated from each other ( Figure 3 represents one such model consistent with our data). We confirmed this result by applying symmetry tests to evaluate the relationships among the Iranian-related lineages, correcting for the effects of Anatolian farmer-related, Andamanese hunter-gatherer-related, and West Siberian hunter-gatherer-related admixture ( Star Methods ). We find that 94% of the resulting trees supported the Iranian-related lineage in the IVC Cline being the first to separate from the other lineages, consistent with our modeling results.

The Iranian-related subtree is shown in green, the Anatolian farmer-related subtree in blue, the southeast Asian-related subtree in red, and mixed populations as pie charts (other fitting topologies all share the feature that the IVC Cline descends from the first-splitting lineage in the subtree of Iranian-related ancestry). The dates of the analyzed populations are shown on the vertical axis and provide minima on the population split dates. The observation that the Iranian-related lineage contributing to the IVC Cline split earlier than Belt Cave at ~10000 BCE and Ganj Dareh at ~8000 BCE is incompatible with the hypothesis that the advent of farming in South Asia after ~7000–6000 BCE was associated with a large-scale eastward migration bringing ancestry from people related to western Zagros mountain farmers or herders across the Iranian plateau to South Asia.
Our evidence that the Iranian-related ancestry in the IVC Cline diverged from lineages leading to ancient Iranian hunter-gatherers, herders, and farmers prior to their ancestors’ separation places constraints on the spread of Iranian-related ancestry across the combined region of the Iranian plateau and South Asia, where it is represented in all ancient and modern genomic data sampled to date. The Belt Cave individual dates to ~10000 BCE, before the advent of farming anywhere in the world, which implies that the split leading to the Iranian-related component in the IVC Cline predates the advent of farming as well ( Figure 3 ). Even if we do not consider the results from the low-coverage Belt Cave individual, our analysis shows that the Iranian-related lineage present in the IVC Cline individuals split before the date of the ~8000 BCE Ganj Dareh individuals, who lived in the Zagros mountains of the Iranian plateau before crop farming began there around ~7000–6000 BCE. Thus, the Iranian-related ancestry in the IVC Cline descends from a different group of hunter-gatherers from the ancestors of the earliest known farmers or herders in the western Iranian plateau. We also highlight a second line of evidence against the hypothesis that eastward migrations of descendants of western Iranian farmers or herders were the source of the Iranian-related ancestry in the IVC Cline . An independent study has shown that all ancient genomes from Neolithic and Copper Age crop farmers of the Iranian plateau harbored Anatolian farmer-related ancestry not present in the earlier herders of the western Zagros ( Narasimhan et al., 2019 ). This includes western Zagros farmers (~59% Anatolian farmer-related ancestry at ~6000 BCE at Hajji Firuz) and eastern Alborsz farmers (~30% Anatolian farmer-related ancestry at ~4000 BCE at Tepe Hissar). That the 12 sampled individuals from the IVC Cline harbored negligible Anatolian farmer-related ancestry thus provides an independent line of evidence (in addition to their deep-splitting Iranian-related lineage that has not been found in any sampled ancient Iranian genomes to date) that they did not descend from groups with ancestry profiles characteristic of all sampled Iranian crop-farmers ( Narasimhan et al., 2019 ). While there is a small proportion of Anatolian farmer-related ancestry in South Asians today, it is consistent with being entirely derived from Steppe pastoralists who carried it in mixed form and who spread into South Asia from ~2000–1500 BCE ( Narasimhan et al., 2019 ).
These findings suggest that in South Asia as in Europe, the advent of farming was not mediated directly by descendants of the world’s first farmers who lived in the fertile crescent. Instead, populations of hunter-gatherers--in Western Anatolia in the case of Europe ( Feldman et al., 2019 ), and in a yet-unsampled location in the case of South Asia--began farming without large-scale movement of people into these regions. This does not mean that movements of people were unimportant in the introduction of farming economies at a later date: for example, ancient DNA studies have documented that the introduction of farming to Europe after ~6500 BCE was mediated by a large-scale expansion of Western Anatolian farmers who descended largely from early hunter-gatherers of western Anatolia ( Feldman et al., 2019 ). It is possible that in an analogous way, an early farming population expanded dramatically within South Asia causing large-scale population turnovers that helped to spread this economy within the region; whether this occurred is still unverified and could be determined through ancient DNA studies from just before and after the farming transitions in South Asia.
Our results also have linguistic implications. One theory for the origins of the now-widespread Indo-European languages in South Asia is the “Anatolian hypothesis,” which posits that the spread of these languages was propelled by movements of people from Anatolia across the Iranian plateau and into South Asia associated with the spread of farming. However, we have shown that the ancient South Asian farmers represented in the IVC Cline had negligible ancestry related to ancient Anatolian farmers, as well as an Iranian-related ancestry component distinct from sampled ancient farmers and herders in Iran. Since language spreads in pre-state societies are often accompanied by large-scale movements of people ( Bellwood, 2013 ) these results argue against the model ( Heggarty, 2019 ) of a trans-Iranian-plateau route for Indo-European language spread into South Asia. However, a natural route for Indo-European languages to have spread into South Asia is from Eastern Europe via Central Asia in the first half of the 2 nd millennium BCE, a chain-of-transmission now documented in detail with ancient DNA. The fact that the Steppe pastoralist ancestry in South Asia matches that in Bronze Age Eastern Europe (but not Western Europe ( de Barros Damgaard et al., 2018 ; Narasimhan et al., 2019 )) provides additional evidence for this theory, as it elegantly explains the distinctive shared distinctive features of Balto-Slavic and Indo-Iranian languages ( Ringe et al., 2002 )
Our analysis of data from one individual from the IVC, in conjunction with 11 previously reported individuals from sites in cultural contact with the IVC, demonstrates the existence of an ancestry gradient that was widespread in farmers in the northwest of peninsular India at the height of the IVC, that had little if any genetic contribution from Steppe pastoralists or western Iranian farmers or herders, and that had a primary impact on the ancestry of later South Asians. While our study is sufficient to demonstrate that this ancestry type was a common feature of the IVC, a single sample--or even the gradient of 12 likely IVC samples we have identified--cannot fully characterize a cosmopolitan ancient civilization. An important direction for future work will be to carry out ancient DNA analysis of additional individuals across the IVC range to obtain a quantitative understanding of how the ancestry of IVC people was distributed, and to characterize other features of its population structure.
STAR Methods
Lead contact and materials availability.
This study did not generate new unique reagents. Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, David Reich ( ude.dravrah.dem.sciteneg@hcier ).

Experimental Model and Subject Details
We attempted to generate genome-wide data from skeletal remains of 61 ancient individuals from the IVC site of Rakhigarhi. Only a single sample yielded enough authentic ancient DNA for analysis: I6113, Rakhigarhi, Haryana, India (n=1). We report the archeological context dates for this burial in Method Details . The skeletal samples from Rakhigarhi were excavated by the archaeological team led by V.S. at the Deccan College Post-Graduate and Research Institute in Pune India and sampled by the ancient DNA group led by N.Ra. at the Birbal Sahni Institute of Palaeosciences in Lucknow India. Analysis using the methods implemented by the ancient DNA group led by D.R. at Harvard Medical School in Boston USA was approved by a Memorandum of Understanding between Deccan College and Harvard Medical School executed in February 2016.
Method Details
Contextual date for individual i6113.
There is insufficient collagen preservation for the human bones at Rakhigarhi cemetery to allow Accelerator Mass Spectrometry radiocarbon dating; multiple attempts showed a carbon-to-nitrogen ratio outside the range appropriate for dating, including five attempts we made specifically on skeletal elements from I6113. However, the cemetery can be securely dated based on archaeological context. First, the only evidence of human occupation of the site is in the Harappan period and hence all the excavated remains are likely to belong to that period. Second, all the characteristic features of the Harappan burial customs and features are present in the cemetery, including a separation from the main habitation area (about 1 kilometer), and typical Harappan artifacts including pots, beads made of semi-precious stones, and bangles of copper, shell or terracotta, all of which are indistinguishable from artifacts found in the main habitation area. As discussed in the text, the main habitation area has 7 radiocarbon dates based on charcoal spanning 2800–2300 BCE, largely falling within the mature IVC ( Shinde et al., 2018 ; Vahia et al., 2016 ). Third, the pottery associated with the burial ( Figure S1 ), appear to be stylistically similar to others made during the mature Harappan period.
Ancient DNA Data Generation
Excel Table Titles and Legends Table S 1 presents details of genetic results on the 251 libraries we generated on 61 distinct samples. To represent I6113, we generated data from 108 libraries (27 double stranded ( Rohland et al., 2018 ) and 81 single stranded ( Meyer et al., 2014 ; Rohland et al., 2015 )), and then filtered out 40 single-stranded libraries (all the libraries from a single extraction) that had extremely low coverage and low levels of the cytosine-to-thymine mismatch to the human reference sequence expected for authentic ancient DNA. For the remaining 68 libraries, we restricted the data to sequences with evidence of characteristic ancient DNA damage in the final nucleotide using PMDtools ( Skoglund et al., 2014 ).
Assessing samples for authenticity of ancient DNA
Based upon the rate of cytosine-to-thymine mismatches to the reference sequence in the final nucleotide of the libraries ( Dabney et al., 2013 ; Korlević et al., 2015 ), we prioritized the individual with relatively high ancient rates of characteristic damage, I6113 for additional library preparation and sequencing ( Excel Table Titles and Legends Table S 1 ).
For I6113 we generated a total of 27 double stranded libraries ( Rohland et al., 2018 ) (1 UDG-treated and 26 not UDG-treated) using powder from both the otic capsule and semicircular canals of the petrous bone, and also generated an additional 81 single stranded libraries (all non-UDG-treated) using powder from the semicircular canal and one of two different extraction procedures ( Meyer et al., 2014 ; Rohland et al., 2015 ). Out of these 108 libraries, nearly all of the 40 made from single-stranded libraries prepared using the extract made with Buffer G ( Korlević et al., 2015 ) had low coverage (<100 targeted SNPs covered) and low damage in the final nucleotide ( Excel Table Titles and Legends Table S 1 ), consistent with the extreme sensitivity of extracts made using this buffer to inhibition especially for single-stranded libraries ( Korlević et al., 2015 ). We therefore removed all libraries prepared from this extract from analysis and proceeded with the remaining 68.
The number of DNA sequences obtained from each library of I6113 was insufficient for assessment of contamination on a per-library basis. We therefore examined the datasets obtained by pooling 208,111 sequences across the 68 libraries for I6113. Examining the number of sequences mapping to the Y chromosome as a fraction of that mapping to both the X and Y, we found a ratio of 0.047. On data for many other ancient individuals subject to ~1.24 million SNP enrichment, we have empirically found that this ratio is less than about 0.03 for uncontaminated libraries from females, and above 0.35 for uncontaminated males. Thus, I6113 has evidence of a mixture of human DNA from both males and females, and thus contamination.
To identify subsets of the molecules that are highly likely to be authentic, we analyzed the fraction of sequences that retained typical signatures of ancient DNA damage based on a characteristic cytosine-to-thymine mismatches to a reference sequence at their ends ( Meyer et al., 2014 ; Skoglund et al., 2014 ), stratified by the lengths of the molecules ( Figure S2A , B ). We carried out this analysis not only for I6113, but also for a previously published ancient DNA sample from Southeast Asia from a similar time period (I4011) comprised of a merge of data from 21 double-stranded libraries ( Lipson et al., 2018 ). The libraries from I6113 have high rate of damage (up to ~50%) indicative of a high proportion of genuine ancient DNA. The rate of damage for I6113 decreases for lengths greater that 40 bp, suggesting that longer molecules are more likely to be contaminated. We further found that sequences that were damaged on one end of the ancient DNA molecules (showing cytosine-to-thymine (C-to-T) mismatches relative to the reference sequence) also had an enhanced chance of damage on the other, as expected if damage restriction enriches for authentic DNA ( Meyer et al., 2014 ) ( Figure S2C ).
To maximize the number of SNPs available for analysis while minimizing contamination, we analyzed multiple subsets of sequences for I6113, restricting to ones with characteristic ancient DNA damage in the final nucleotides. The resulting dataset contains sequences covering 31,760 SNPs at least once. Its ratio of Y chromosome sequences to X+Y chromosome sequences is 0.026, consistent with being an uncontaminated female (and the anthropological determination).
Autosomal Contamination Estimates
We estimated contamination using an algorithm based on breakdown of linkage disequilibrium ( Posth et al., 2018 ). This software measures contamination levels by comparing the haplotype distribution of a tested sample to the haplotype distribution of an external reference panel. We used Sri Lankan Tamils sampled from the United Kingdom (STU) from the 1000 Genomes Project ( Auton et al., 2015 ) as the reference panel. The algorithm was run in the usually conservative “uncorrected” mode to attain maximal power.
Quantification and Statistical Analysis
Admixture clustering analysis.
We pruned the data using PLINK2 to retain only sites for which at least 70% of individuals had a non-missing genotype ( Chang et al., 2015 ). We then ran ADMIXTURE ( Alexander et al., 2009 ) with 10 replicates and report the replicate with the highest likelihood. In Figure 2 , we show the results for clustering using K=6 components.
f -statistics
We computed f -statistics using the packages in ADMIXTOOLS ( Patterson et al., 2012 ). To test for admixture we ran f 3 -statistics using the inbreed:YES parameter with an ancient population as a target. To estimate the ancestry proportion for a test population given a set of source populations and a set of outgroups, we used the qpAdm methodology ( 18 ) in ADMIXTOOLS.
Hierarchical modeling
To model a given sample as part of an established genetic cline determined by a set of other populations, we used an approach described in ( Narasimhan et al., 2019 ) (the Supplementary Materials of ( Narasimhan et al., 2019 ) gives the full details). We begin by obtaining ancestry proportions for a set of samples on a genetic cline, and jointly model these in a single generative model taking advantage of the fact that the proportions for the three ancestral sources must sum to 1. We estimate the mean and covariance of these sources using a bivariate normal distribution via maximum likelihood. We evaluate if the test population can be fit as deriving from the same original three sources as those we just modeled on the genetic cline using qpAdm , and if there is a fit, evaluate if the observed ratios of the ancestry proportions of the test population fit with the expected values from the generative model established by the cline. We compute a p-value based on the empirically determined covariance matrices.
Determination of the phylogeny of Iranian-related ancestry
We wished to examine the relationship of the Iranian-related ancestry present in the IVC Cline to that of ancient Iranian plateau individual reported in the ancient DNA literature.
We first focused on a set of populations chosen to represent a diverse group of early hunter-gatherers and farmers from across the geographic area of present day Iran. Our approach was to identify a set of allowable phylogenies and then, based on the known dates of the samples, to make inferences on minimum split times between lineages.
The individuals or groups of individuals we examined were:
- Belt_Cave_M (BC) (n=1) – A Mesolithic individual from the Alborz mountains of Central Iran. Due to the evidence of contamination in the data from this individual, we used a damage restricted version of the sample resulting in 30,722 SNPs.
- Ganj_Dareh_N (GD) (n=8) – Early goat herders from the Zagros Mountains of western Iran. The highest coverage individual has data from 938,523 SNPs.
- Hajji_Firuz_C (HF) (n=5) – Late Neolithic and early Copper Age individuals from the Zagros Mountains of Western Iran. The highest coverage individual has data from 916,581 SNPs.
- Tepe_Hissar_C (TH) (n=12) – Copper Age individuals from the Central Iranian Plateau. The highest coverage individual has data from 745,066 SNPs.
- Indus_Periphery_West (IP) (n=1) – Member of the IVC Cline which includes the Rakhigarhi individual I6113. We represent the Iranian-related ancestry in this cline with I8728, the individual with the highest Iranian-related ancestry and also the highest coverage on this cline with data from 657,401 SNPs.
As documented in ref. ( Narasimhan et al., 2019 ), the Hajji Firuz and Tepe Hissar pools of individuals have evidence of admixture related to Anatolian farmers while the Indus Periphery individuals (of which we show several including the individual with the highest West Eurasian-related ancestry I8728) have significant proportions of ancestry related to southeast Asian hunter-gatherers.
Building scaffolds of all possible topologies of Iranian-related ancestry
We were interested in understanding the relationship of the Iranian-related component of the ancestry of these 5 populations, treating the non-Iranian-related components such as the Anatolian farmer-related ancestry as nuisances that we need to model out assuming a topology in which the lineages lead to Tepe Hissar (PTA) and Hajji Firuz (PHA) formed a separate clade from Anatolian farmers (in the next sub-section we show that our results are robust to the choice of the topology relating the Anatolian farmer-related lineages).
There are 3 distinct topologies according to which these 5 populations could be related, which we call “Serial Founder” ( Figure S3A ), “Single Outgroup” ( Figure S3B ), and “Two Clades” ( Figure S3C ). Within these topologies where are multiple permutations for how the 5 individual populations could relate, depending on how the 5 Iranian-related populations fit into “slots” on the topology.
We used qpWave ( Patterson et al., 2012 ) to evaluate all 120 possible ways for the 5 populations to be grafted onto each of the open “Slots” or positions, taking care to account for the correct admixing source for the populations that were admixed. In some topologies the assignment of populations to the slots did not alter the graph topology (when two populations were a clade with respect to the others. Therefore, there were only 30, 15 and 60 different models that were unique for the Serial Founder, Single Outgroup and Two Clades phylogenies respectively, though in our results we show all 120 possibilities for the assignment of a “Slot” to a population.
Methodology for model selection of admixture graphs
In the previous section we described the assignment of populations to “Slots” and the creation of a large number of admixture graphs.
For each admixture graph produced there are two metrics that we used to evaluate fit. The first is a list of residuals above a particular Z-score and the second is a score for the weighted error for the fitted statistics based on the graph in comparison to the empirically observed statistics, S ( G ) = −1/2( g − f )′ Q −1 ( g − f ). Here f is the vector of observed f- statistics and g is the corresponding vector of statistics fit on the graph with the specified topology, and Q is an estimated covariance matrix determined empirically ( Patterson et al., 2012 ).
As a first screen for a working model, we begin by selecting models that have their largest residual with an absolute Z-score below 3. This is a standard approach in the literature and while this may provide a practical threshold for rejecting models, this alone is not sufficient to adjudicate between two models whose worst fitting residuals are both close to the threshold. We wanted to get an idea of how similar two models were with respect to their statistical likelihood. If our admixture graph models were nested within one another, we could do this using a Likelihood Ratio Test as described in ( Lipson and Reich, 2017 ). In this approach, the log-likelihood of two models, one involving admixture from a certain population and one without were compared and their difference can then be compared using a chi-squared test.
However this approach cannot be applied in the present setting as the models we are testing are not nested within one another and the number of parameters of all of the models is the same. To enable us to examine the level of support one particular model has when compared to another we use Akaike Information Criterion (AIC) as a measure of model fit. Since the number of parameters between two models remains the same, the actual computed score remains the same whether we use AIC or a Bayesian Information Criterion (BIC).
We used a set of guidelines outlined in ( Burnham and Anderson, 2004 ) for performing model selection using AIC. Specifically we computed δ i = AIC i − AIC min where AIC i is the AIC of the i -th model, and AIC min is the lowest AIC obtained amongst all the models we tested across all topologies (SO, SF and TC). The models can then be compared using the following guidelines from ( Burnham and Anderson, 2004 ):
- δ i ≤ 2, the i -th model is nearly as plausible as the best fitting model;
- 2 < δ i ≤ 4, the i- th model is consistent with the data but considerably less probable than the best fitting model;
- 4 < δ i ≤ 7, the i -th model is much less likely than the best fitting model;
- 7 < δ i , the i -th model has essentially no support.
Based on this published set of criteria we chose to accept all models for which the difference in AIC was <4.
Among the samples we analyzed was a Mesolithic individual from Central Iran, Belt_Cave_M , which after restricting to damaged sequences (to address the evidence of contamination in this individual) reflects data from just 30,722 autosomal SNPs, and thus co-modeling with this individual restricts the number of SNPs available for admixture graph fitting. To address this, we repeated the admixture graph fitting removing this particular individual which improved our SNP coverage by more than 10 fold, allowing us to remove models that worked simply because of a lack of data and ensuring that the fit of a particular admixture graph was not due to our inability to reject it at lower coverage. As a further criterion for model selection, we restricted to the intersection between the fitting models analyzed with and without the Belt Cave individual.
Results from the model selection of tested admixture graphs
We report the successful graph topology results of our admixture graphs in Table S3 .
In Table S3A , we observe that all working models exclude the “Two Clades” topology, regardless of how populations get assigned to “slots”. We also observe that all fitting populations have Indus_Periphery_West or Hajji_Firuz as an outgroup with respect to all other groups at the AIC < 4 threshold, with only models with Indus_Periphery_West if we use AIC<2.
Robustness to altering the topology of the non-Iranian-related admixing sources
We explored if modifying the ordering of both the South Asian hunter-gatherer-related components ( Table S 3B ) and Anatolian farmer-related admixture events ( Table S 3C ) change our inferences and found that they did not except in one notable way. Previously our model selection criterion had not been successful at distinguishing the earliest diverging of the Iranian-related populations. i.e. either Indus_Periphery_West or Hajji_Firuz_C , but under a different topology of the Anatolian farmer-related component in Hajji_Firuz_C we see that models with Indus_Periphery_West as the earliest diverging split are strongly supported over the other working models.
The inference is robust to treating all of the test populations as admixed
As another way of perturbing the resulting graphs, we chose to allow all the five populations to be admixed with all source populations thereby allowing a much freer model. We find that our results showing that the Indus_Periphery_West being the first to split are robust to this perturbation ( Table S 3D ).
Taken together, this analysis shows a clear branching structure which involves the Indus_Periphery_West ancestry as the first to split, followed by the others which are not distinguishable. There are two marginally fitting models in which Hajji_Firuz is the first to split ( Table S 3A ), but even if these models are correct they do not change the inference that the Iranian-related ancestry in Indus_Periphery_West split from the lineages leading to those in Belt Cave, Tepe Hissar, and Ganj Dareh before they separated from each other, which is the only inference we need for our main conclusions.
Alternative approaches to determining phylogeny
The major confounder when inferring trees and examining their topology as determined by shared drift amongsdifferent populations is admixture. In the analyses above based on testing of admixture graphs, we dealt with this by modeling known admixtures into populations, and showed that the changing of the topology of the admixing sources does not affect the inference we obtain about the internal phylogeny of the Iranian related component of the ancestry of our test populations. As an alternative approach to exploring these issues, we obtained unbiased estimates of the allele frequencies for the Iranian-related component in each of the samples by subtracting the expectation from the admixing sources, and then performed symmetry tests to reconstruct the phylogenetic relationships.
Prior to implementing this procedure on real data, we began by confirming that if the relevant admixing source populations or populations related to those source populations were available and the proportion of their admixture known, then it was possible to recover the internal phylogeny of the populations even though there is significant admixture present in the data.
To do this we simulated the phylogeny described in Figure S4 using the msprime coalescent simulator ( Kelleher et al., 2016 ). We used standard mutation and recombination rates and sampled 1 million positions in 10 individuals from each population. We converted these to haploid genotypes by random sampling. We were interested in whether we could recover the internal phylogeny of the pp5 node. The choice of this particular topology and set of admixing populations mirrors the structure of the admixture graph that we think may be a reasonable match to our real data.
In the first step of the process, we computed the allele frequencies per SNP for the populations for which we were interested in obtaining a phylogeny, namely 3, 4, 5 and 6. We then subtracted the relevant allele frequencies of the admixing populations which were known in this setting. For example, we subtracted the allele frequency of population 8 from population 6, weighted by the admixture proportion 50%. We then computed all statistics of the form f 4 ( 0,A,B,Test ), where A, B and Test could be any of the populations 3, 4, 5 and 6. In Table S 3E , we show that for samples without admixture correction there are no simple trees that are compatible with the data and it is not possible to uncover the internal phylogeny of these populations (at the |Z|>3 level).
However, after subtracting the allele frequencies ( Table S 3F ), it is possible to infer the internal phylogeny of the graph under our threshold. This suggests that if we account for the correct admixing population as well as the proportion of admixture, it is possible to recover the phylogeny of a set of populations even though they might be admixed even to levels of 50% as was the case in simulations. In the next section, we apply this procedure to the admixture graph that we constructed using the real data.
Accounting for admixture at the genotype level on real data
We began by examining if our inference procedure that we used on the simulated data could be applied as an additional validation of our best fitting admixture graph. To do this, we computed the allele frequencies of the Iranian-related populations, Ganj_Dareh_N , Hajji_Firuz_C , Tepe_Hissar_C and Indus_Periphery_West . We dropped Belt_Cave_M as it was too low in coverage to produce meaningful results. We then examined the inferred admixture proportions of the non-Iranian-related ancestry in these populations. To do this we utilized the qpAdm methodology for distal populations that was developed in ( Narasimhan et al., 2019 ), and tested the populations that were required for admixture graph fitting, AHG , and Anatolia_N as sources for all of the populations. This produced point estimates that were in line with our admixture graph fits for the relevant admixture proportions as well as a covariance matrix measuring uncertainty.
To account for uncertainty in this procedure, we carried out this procedure sampled 1000 times from the point estimates and covariance matrix of admixture proportions and produced 1000 samples. For each of these samples we subtracted the allele frequencies of the AHG- and Anatolia_N -related ancestries and computed all possible triplets of f 4 -statistics as we had done for the simulated data. We computed f 4 -statistics using a |Z|>3 threshold to determine whether there continued to remain significant evidence of admixture relating the populations. Unlike with the simulated data where we knew a priori the exact mixing proportions and admixing sources, the uncertainty in the admixture proportion resulted in reduced power. We observed 465 cases where the inferred tree was (IP,(GD,(HF,TH))), 29 cases where the inferred tree was (IP,(HF,(GD,TH))), and 506 cases where a single tree determination could not be made due to the presence of additional significant admixture events between the populations. These results suggest that using this procedure we produce only two viable tree topologies, both of which involve Indus_Periphery_West as the population splitting first, mirroring the topology produced using the admixture graph methodology.
Data and Code Availability
Additional resources.
Supplemental Data include an Excel spreadsheet detailing all ancient samples for which attempts were made to extract ancient DNA data and excel spreadsheets with other statistics detailed in the paper.
Key Resources Table
- The sample was from a population that is the largest source of ancestry for S Asians
- Iranian-related ancestry in S Asia split from Iranian plateau lineages >12000 yr ago
- First farmers of the Fertile Crescent contributed little to no ancestry to S Asians
Supplementary Material
Figure S1 . Ceramics in the Grave of Individual I6113/RGR7.3, BR-01, HS-02, Related to Figure 1 and STAR Methods. (A) Broken Goblet from Harappan level placed at the top of the head. (B) Complete specimen of a Red Slipped ware medium-sized globular pot from the Harappan level placed at the top of the head. (C) Central Column of a Stand from the Harappan level that appears to have been burned, placed at the top of the head. (D) Broken specimen of Red Slipped ware medium size globular pot from the Harappan level placed at the top of the head. There are lines as well as indentations on the upper right side, just below the rim. (E) Broken specimen of a Red Slipped ware medium-sized globular pot from Harappan level placed at the top of the head. There are a series of linear marks suggesting that these pots were wheel-thrown. Additional indication of this is seen in the interior of the vessel. (F) Complete specimen of a Red Slipped ware beaker from the Harappan level placed at the top of the head. (G) Complete specimen of Red Slipped ware large vessel from the Harappan level placed at the top of the head. The distinct rim suggests that this was made separately and then attached to the pot. (H) Broken stand of a dish on stand from the Harappan level placed at the top of the head. (I) Complete beaker from the Harappan level placed near the head. (J) Complete Goblet from the Harappan level placed at the top of the head. (K) Broken black painted Goblet from the Harappan level placed at the top of the head. (L) Complete beaker from the Harappan level placed near the right leg.
Figure S2. Quality control to identify a subset of authentic sequences, Related to STAR Methods. (A) The distribution of the number of sequences of a given length for two different samples - I6113 from Rakhigarhi, and I4011 which is previously reported data of a similar chronological age from Myanmar. For I6113, sequence length distributions are shown separately for pools of the double and single stranded libraries. (B) The fraction of sequences showing characteristic ancient DNA damage as a function of sequence length for I6113 and I4011. For I6113, sequence length distributions are shown separately for double and single stranded libraries. (C) Frequency of C-to-T substitutions for both ends of the sequences as a function of distance from the end for I6113, after merging all libraries. The profile shows damage patterns characteristic of authentic ancient DNA. Restricting to sequences with C-to-T substitutions on one end of a fragment result in an increase in the damage rate on the other end.
Figure S3. Phylogenies of Iranian-related populations tested for fits to the data, Related to STAR Methods. (A) “Serial Founder” model: A first population splits, then a second, then a third, then a fourth and fifth. (B) “Single Outgroup” model: A single population splits. Thereafter two pairs of populations diverge from a common source. (C) “Two Clades” model: The 5 populations split into two groups, one with 3 populations in a clade and the second with 2 populations in a clade.
Figure S4. Simulated phylogeny, Related to STAR Methods. This was the phylogeny used to test our inference procedure.
Table S 1, Related to STAR Methods. Detailed information for the 251 libraries generated for this study
Table S 2, Related to STAR Methods. Summary information for each of the 61 individuals screened for this study
Table S 3a-d, Related to STAR Methods. Z-scores, likelihoods and AIC of difference from best performing model for the admixture graph topologies shown in Figure S3 along with results from simulations of admixture graphs and all possible f-statistics before and after correcting for admixture.
Acknowledgements
We thank Richard Meadow and several anonymous reviewers for critical comments on the manuscript and suggestions for improvements, and Anna Koutoulas and the Genomics Platform at the Broad Institute for sequencing support. We thank Elena Essel and Sarah Nagel for help in the laboratory. K.T. was supported by NCP fund (MLP0117) of the Council of Scientific and Industrial Research (CSIR), Government of India. D.R. is an Investigator of the Howard Hughes Medical Institute and his ancient DNA laboratory work was supported by National Science Foundation HOMINID grant BCS-1032255, by National Institutes of Health grant GM100233, by an Allen Discovery Center grant, and by grant 61220 from the John Templeton Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
- Alexander DH, Novembre J, and Lange K (2009). Fast model-based estimation of ancestry in unrelated individuals . Genome research 19 , 1655–1664. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, Flicek P, et al. (2015). A global reference for human genetic variation . Nature 526 , 68–74. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Behar DM, van Oven M, Rosset S, Metspalu M, Loogväli E-L, Silva NM, Kivisild T, Torroni A, and Villems R (2012). A Copernican reassessment of the human mitochondrial DNA tree from its root . American journal of human genetics 90 , 675–684. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Bellwood PS (2013). 11 Human migrations and the histories of major language families In The Encyclopedia of Global Human Migration , Ness I, ed. (Chichester, UK: Wiley-Blackwell; ), pp. 87–95. [ Google Scholar ]
- Broushaki F, Thomas MG, Link V, López S, van Dorp L, Kirsanow K, Hofmanová Z, Diekmann Y, Cassidy LM, Díez-Del-Molino D, et al. (2016). Early Neolithic genomes from the eastern Fertile Crescent . Science (New York, NY) 353 , 499–503. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Burnham KP, and Anderson DR (2004). Multimodel Inference: Understanding AIC and BIC in Model Selection . Sociological Methods & Research 33 , 261–304. [ Google Scholar ]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, and Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets . GigaScience 4 , 7. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Dabney J, Knapp M, Glocke I, Gansauge M-T, Weihmann A, Nickel B, Valdiosera C, García N, Pääbo S, Arsuaga J-L, et al. (2013). Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments . Proceedings of the National Academy of Sciences of the United States of America 110 , 15758–15763. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- de Barros Damgaard P, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, et al. (2018). The first horse herders and the impact of early Bronze Age steppe expansions into Asia . Science (New York, NY) 360 , eaar7711. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Feldman M, Fernández-Domínguez E, Reynolds L, Baird D, Pearson J, Hershkovitz I, May H, Goring-Morris N, Benz M, Gresky J, et al. (2019). Late Pleistocene human genome suggests a local origin for the first farmers of central Anatolia . Nature Communications 10 , 1218. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. (2015). An early modern human from Romania with a recent Neanderthal ancestor . Nature 524 , 216–219. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. (2015). Massive migration from the steppe was a source for Indo-European languages in Europe . Nature 522 , 207–211. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Heggarty P (2019). Prehistory through language and archaeology . In (Routledge; ), pp. 616–644. [ Google Scholar ]
- Kelleher J, Etheridge AM, and McVean G (2016). Efficient Coalescent Simulation and Genealogical Analysis for Large Sample Sizes . PLOS Computational Biology 12 , e1004842. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Korlević P, Gerber T, Gansauge M-T, Hajdinjak M, Nagel S, Aximu-Petri A, and Meyer M (2015). Reducing microbial and human contamination in DNA extractions from ancient bones and teeth . BioTechniques 59 , 87–93. [ PubMed ] [ Google Scholar ]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. (2016). Genomic insights into the origin of farming in the ancient Near East . Nature 536 , 419–424. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Li H, and Durbin R (2010). Fast and accurate long-read alignment with Burrows–Wheeler transform . Bioinformatics 26 , 589–595. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Lipson M, Cheronet O, Mallick S, Rohland N, Oxenham M, Pietrusewsky M, Pryce TO, Willis A, Matsumura H, Buckley H, et al. (2018). Ancient genomes document multiple waves of migration in Southeast Asian prehistory . Science 361 , 92–95. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Lipson M, and Reich D (2017). A Working Model of the Deep Relationships of Diverse Modern Human Genetic Lineages Outside of Africa . Molecular biology and evolution 34 , 889–902. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. (2015). Genome-wide patterns of selection in 230 ancient Eurasians . Nature 528 , 499–503. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Meyer M, Fu Q, Aximu-Petri A, Glocke I, Nickel B, Arsuaga J-L, Martínez I, Gracia A, de Castro JMB, Carbonell E, et al. (2014). A mitochondrial genome sequence of a hominin from Sima de los Huesos . Nature 505 , 403–406. [ PubMed ] [ Google Scholar ]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. (2012). A high-coverage genome sequence from an archaic Denisovan individual . Science (New York, NY) 338 , 222–226. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Mughal MR (1990). Further evidence of the Early Harappan Culture in the Greater Indus Valley . South Asian Studies 6 , 175–2000. [ Google Scholar ]
- Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, et al. (2019). The formation of human populations in South and Central Asia . Science 365 , eaat7487. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, and Reich D (2012). Ancient admixture in human history . Genetics 192 , 1065–1093. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Possehl GL (1982). The Harappan Civilization: A contemporary perspective In Harappan Civilization (Oxford University Press; ), pp. 16–28. [ Google Scholar ]
- Possehl GL (1990). Revolution in the Urban Revolution: The Emergence of Indus Urbanization . In Annual Review of Anthropology (Annual Reviews) , pp. 261–282. [ Google Scholar ]
- Posth C, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, Rohland N, Nägele K, Adamski N, Bertolini E, et al. (2018). Reconstructing the Deep Population History of Central and South America . Cell 175 , 1185–1197.e1122. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Ringe D, Warnow T, and Taylor A (2002). Indo-European and Computational Cladistics . Transactions of the Philological Society 100 , 59–129. [ Google Scholar ]
- Rohland N, Glocke I, Aximu-Petri A, and Meyer M (2018). Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing . Nature Protocols 13 , 2447–2461. [ PubMed ] [ Google Scholar ]
- Rohland N, Harney E, Mallick S, Nordenfelt S, and Reich D (2015). Partial uracil-DNA-glycosylase treatment for screening of ancient DNA . Philosophical Transactions of the Royal Society B: Biological Sciences 370 , 20130624. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Shaffer JG, and Lichtenstein DA (1989). Ethnicity and change in the Indus Valley cultural tradition In Wisconsin Archeological Reports , Kenoyer JM, ed., pp. 117–126. [ Google Scholar ]
- Shinde VS, Kim YJ, Woo EJ, Jadhav N, Waghmare P, Yadav Y, Munshi A, Chatterjee M, Panyam A, Hong JH, et al. (2018). Archaeological and anthropological studies on the Harappan cemetery of Rakhigarhi , India. PLOS ONE 13 , e0192299. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, and Jakobsson M (2014). Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal . Proceedings of the National Academy of Sciences 111 , 2229–2234. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- Thapar R (1979). The Mosaic of the Indus Civilization Beyond the Indus Valley . In International Conference on Mohenjo-daro, Karachi. [ Google Scholar ]
- Vahia MN, Kumar P, Bhogale A, Kothari DC, Chopra S, Shinde VS, Jadhav N, and Shastri R (2016). Radiocarbon dating of charcoal samples from Rakhigarhi using AMS . Current Science 111 . [ Google Scholar ]
DNA analysis of Rakhigarhi remains challenges Aryan invasion theory
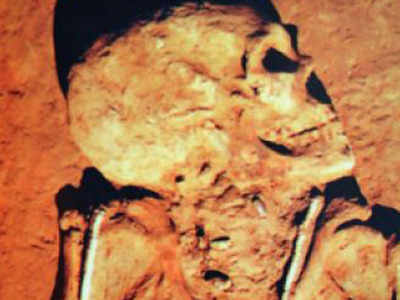
Visual Stories
Rakhigarhi skeleton DNA: Indus Valley people not Rig-Vedic Aryans
New research has debunked purist far-right wing arguments about the settling of the Indian subcontinent and proved the role of migration and cross-cultural exchange in the growth of civilization
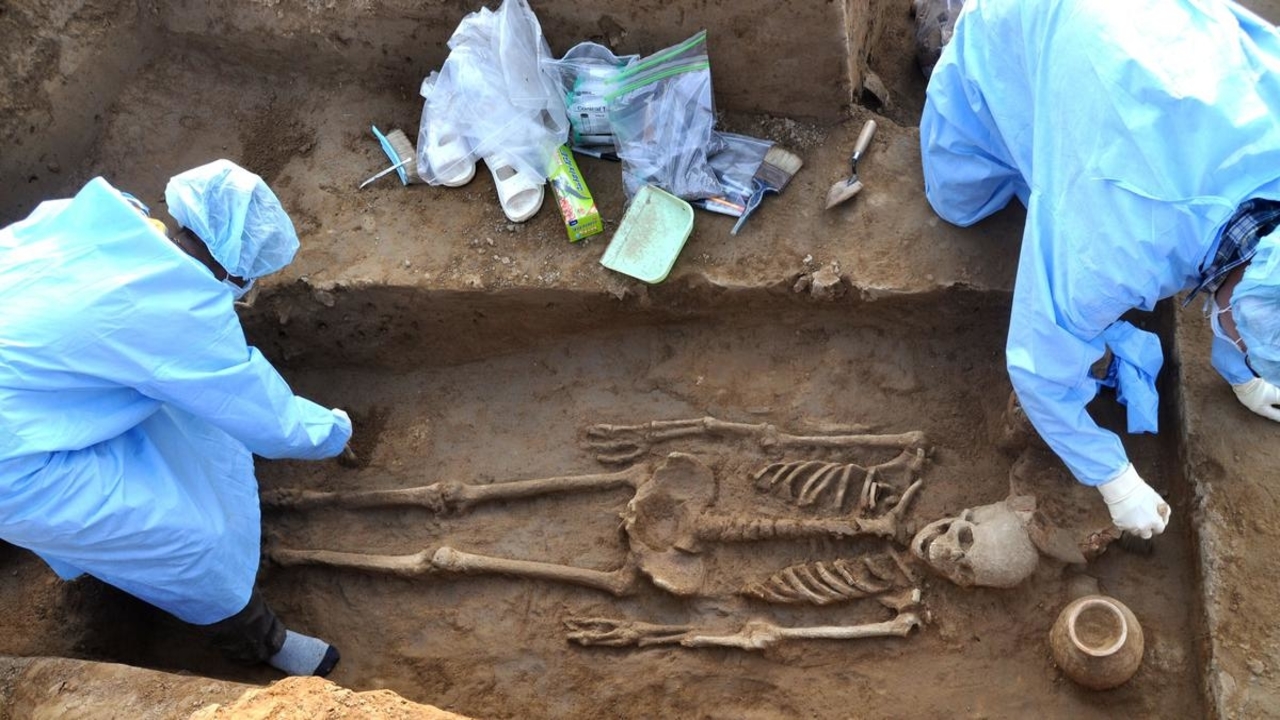
- Ground Reports
- 50-Word Edit
- National Interest
- Campus Voice
- Security Code
- Off The Cuff
- Democracy Wall
- Around Town
- PastForward
- In Pictures
- Last Laughs
- ThePrint Essential


New Delhi: The study of DNA samples of the skeletons found in Rakhigarhi, an Indus Valley Civilisation site in Haryana, has found no traces of the R1a1 gene or Central Asian ‘steppe’ genes, loosely termed as the ‘Aryan gene’.
The study — titled ‘ An ancient Harappan genome lacks ancestry from Steppe pastoralists or Iranian farmers ’ — examined the DNA of the skeletal remains of an individual in Rakhigarhi dating back to around 2500 BC, which was part of the ‘mature Harappan civilisation’ or the Indus Valley Civilisation (IVC).
“The population has no detectable ancestry from Steppe pastoralists or from Anatolian and Iranian farmers, suggesting farming in South Asia arose from local foragers rather than from large-scale migration from the West,” said the study published Friday.
The Central Asian ‘steppe’ gene is found in much of the Indian population today.
“These individuals (in Rakhigarhi) had little of any Steppe pastoralist-derived ancestry, showing that it was not ubiquitous in north-west South Asia during the IVC as it is today,” according to the study.
It added, “While there is a small proportion of Anatolian farmer-related ancestry in South Asians today, it is consistent with being entirely derived from Steppe pastoralists who carried it in mixed form and who spread into South Asia from 2000–1500 BCE.”
The study, led by archaeologist Vasant Shinde, concluded: “Our analysis of data from one individual from the IVC…demonstrates the existence of an ancestry gradient that was widespread in farmers to the northwest of peninsular India at the height of the IVC, that had little if any genetic contribution from Steppe pastoralists or western Iranian farmers or herders, and that had a primary impact on the ancestry of later South Asians.”
A disclaimer
However, towards the end of the study, there is a disclaimer, stating, “While our study is sufficient to demonstrate that this ancestry profile was a common feature of the IVC, a single sample — or even the gradient of 12 likely IVC samples we have identified — cannot fully characterise a cosmo-politan ancient civilisation.”
The Aryan invasion theory is said to be the brainchild of India’s former colonial rulers, who peddled the idea that members of the country’s high castes were descendants of Aryan invaders from Central Asia who are also the forebears of the Europeans.
Some Hindu groups subsequently argued that Aryans were not invaders at all, but native to the land, and that these indigenous people developed Vedic Hinduism. The invasion theory suggests that Vedic Hinduism was developed by European migrants, and came after the Indus Valley civilisation.
Also read: New study shows climate change caused decline of flourishing Indus Valley Civilisation
‘Movement of Central Asians towards Indus Valley not invasion’
Addressing a press conference discussing the findings of the study, Vasant Shinde said much of the development associated with the “foreigners” was brought about by the Indus Valley Civilisation.
“Earlier, it was thought that development only started with the movement of those from Central Asia and West Asia towards the Indus Valley Civilisation. But that is incorrect. All the development was done by indigenous people,” Shinde said.
However, the authors of the study were wary of calling the movement of Central Asians an “invasion”.
Dr Niraj Rai, genetic researcher headed the study with Shinde, said there still isn’t enough evidence to call the movement an “invasion”.
Explaining how contemporary South Asians could have steppe genes, when it wasn’t found in the Rakhigarhi DNA samples, Rai said, “There is a difference between migration and movement. There was certainly some mixing and assimilation, but we can’t call that an invasion,” said Rai.
Draft of the study had created furore
A draft of the Rakhigarhi study published last year had created a wave of discussion when it was reported that the skeletons lacked the R1a1 gene.
A report suggested that the findings of the study reinforced the Aryan invasion theory, because no Central Asian ancestry could be found in the DNA sample excavated.
“We did not find any Central Asian ancestry in the DNA sample. This suggests that the Rakhigarhi residents hadn’t mixed with the Central Asians till then,” Niraj Rai had told ThePrint.
However, Shinde, who led the 2015 excavation in Rakhigarhi, had debunked reports that said the study confirmed the Aryan invasion.
“Plenty of people believe the Aryans came from Central Asia. But we have no evidence of that,” Shinde had said .
Also read: Archeologist who found 4,500-yr-old skeletons in Haryana doesn’t buy Aryan invasion theory
Subscribe to our channels on YouTube , Telegram & WhatsApp
Support Our Journalism
India needs fair, non-hyphenated and questioning journalism, packed with on-ground reporting. ThePrint – with exceptional reporters, columnists and editors – is doing just that.
Sustaining this needs support from wonderful readers like you.
Whether you live in India or overseas, you can take a paid subscription by clicking here .
- Indus Valley Civilisation
41 COMMENTS
The R1 haplogroup is a Y chromosome haplogroup and this is a woman. The findings have nothing to do with the R1 haplogroup. Please correct that.
The genome model didn’t come from the woman alone. There were 11 more individuals. Some of them men.
So is India the origin of mankind?
Despite the ludicrous claims made by Shinde , one only needs to read the original article to see that this study is the strongest ever evidence yet that an Aryan Migration DID take place. Shinde / Rai can keep discussing the difference between migration and invasion, but it is clear that the composers of the Rig veda had origins outside India.
The study says IVC people had no aryan genes. This is correct. We also know , through multiple previous genetic studies that modern Indians carry the aryan gene, especially upper-caste Hindus in North India. So where did this gene come from ? Not from the IVC because the IVC does not have aryan genes. So the only way it could have come is from Flipkart or Amazon !! And if that is not acceptable, the ONLY remaining option is Aryan migration.
On page 7, the study says <<> This in simple language means Aryan migration..
Why is Shinde inviting ridicule upon himself ? Or is he he under the same pressures that delayed the report by 2 years. ?
The quote from the report has been left out. Repeating here On page 7, the paper says <> This in simple language means Aryan migration.
You’re too fake to be believed, and apparently the cell websites requires paid subscription so normal people can’t freely visit it. For “Aryan” genes in Modern Indians: Dr.Shinde did mention that modern Indian do have little steppe DNA, and it is due to Steppe migration between 2500-1500BCE.
What’s new in this study? Everyone knew that during the time of IVC ,there were no migration of steppe people. They came later during 1900-1600 bce.
Birth is a simple event of chance.The mother’s ovum getting fertilised by one of a billion sperms probabilistically ensuring equality of genders. That I was born in India into some religion or cast is but a product of this chance. My effort to claim a divine favour and link to some racial superiority is simply a hoax of egoistic and divisive identity.This led to us keeping our fellow humans, generation after generation, at sub human levels as shudras etc claiming divine knowledge of Vedas.As Indians we should celebrate that today we represent a fine mosaic of early humans attracted to this land for its geography so well suited to agriculture that built all the civilisations. Certainly as Aryans came to this fertile land of Indo gangetic plains and developed these for pastoral living,our”adivasi”brotheren to forest living of central India. Check their DNA ! All the DNA studies should be only to understand the evolution of human societies subsequent to “African Mother”. Except for propagating racism, as its hidden agenda, and parochialism the debate adds little to knowledge per se.In the 21st century it id time to talk humanism.
Very well said <>
<> I could not agree more. Unfortunately , those who who oppose the aryan migration as a matter of “national honour” are the same people who originated and perpetuated the caste system.
North Indian I. E. Punjabi dalit have r1a dna, so upper caste hindus are not outsider only, steppe people mixed with all harrapan, and then create heriarchy.
Muslims are dancing naked here with these studies… Obviously there are unable to comprehend what’s being mentioned in report
Everyone know ivc hasn’t any relation with Indo Aryan.. Indo Aryan migration happened after ivc period or in decline period if ivc… So the study establish no indigenous Aryan.. Aryans came from out side
It simply indicates that Aryans were not native to this region since ‘Indua Valley Civilization’ was first in this region. The Aryans came much later as in this region and tried to impose their ideology. This is now a concretely proven fact.
What needs to be studied is its contemporary civilization found in Sanauli Village of Uttar Pradesh. They now found over 100 coffins there. Along with the coffins, they found swords, daggers, shields, one of the oldest helmets on earth, and a chariot. This was an amazing find. They should do DNA studies on those skeleton remains found. That will help with many of the mysterious regarding the DNA of Indians.
Print didn’t say to whom did 2500 BC skeleton DNA belongs to or say indus valley had three development stages nor does it explains central Asian immigrants became Europeans. 2500 BC remain belongs to tribe in Tamil Nadu, where they had similar script to early indus. 17% male DNA of current North Indian male have Iranian and steppe DNA (immigrants where mostly male) And influence of old pagan religions on India.
Haven’t read the original article . Comments : 1 . Evidence to prove that the skeleton (s) was 2500yrs old ? 2 . On basis of anthropometric characteristics , Indian population appears to have resemblance with Caucasians (NW) , Mangolians (NE) & Negros (South). 3. Cultural practices, language, scriptures , architecture, food habits of different regions of the subcontinent have resemblance with Mayan civilization , Middle Eastern civilizations, North African civilizations, Chinese civilizations. 4. Indian people ( exception of some indigenous Tribes ) have immigrated from various parts of the world. Some researchers have claimed that Kandha Tribe in Kandhamal District of Odisha are not indigenous tribes but immigrated from Africa. 5. This particular paper doesn’t look like a Genetic Study aiming at the origins of Indian people. But written with a motive keeping in mind the present days of social unrest between castes , religious faiths ; political uprising of a group with a religious & ideological belief . This is an attempt to strengthen the argument of the latter group of people. Such papers are a ridicule of scientific research.
Exactly..but I don’t know why Hindi newspapers of my area are citing this study to establish Indo European rigvedic people were natives.
One thing being hidden to public is that Dravidians or Tamils DNA link to the IVC instead being quoted as South Asian.
We fail again and again to understand the game plan of western civilizations. They came to India just 3-4 centuries ago and have the arrogance of calling people living for few thousands of years in India as invaders!!
Firstly, they create and foist their own agenda of Aryan invasion on India. On their own they divided India into Aryan & non-Aryan. Non-Aryans were called Dravidians although there existed none such using their favourite, skin-color. Then created suspicions and hate by ascribing their theory of Aryan invasion theory on caste lines.
Secondly in modern times, as science and archeology developed, selective use of evidence were used to justify their false theories. In-house academicians were recruited to give an air of cridibility and to propagate.
Thirdly, when new evidences are researched which negates their agenda, these western people in certain commentaries (The Atlantic, today) allege that Indian right-wing people may mis-use it. Although we have all along been saying that Aryan Invasion theory has been wrong, we are castigated for supporting and propagating the truth.
Divide & Rule !!
From the actual Vasant Shinde research paper:
“a natural route for Indo-European languages to have spread into South Asia is from Eastern Europe via Central Asia in the first half of the 2nd millennium BCE, a chain of transmission that did occur as has been documented in detail with ancient DNA. The fact that the Steppe pastoralist ancestry in South Asia matches that in Bronze Age Eastern Europe (but not Western Europe [de Barros Damgaard et al., 2018, Narasimhan et al., 2019]) provides additional evidence for this theory, as it elegantly explains the shared distinctive features of Balto-Slavic and Indo-Iranian languages (Ringe et al., 2002).”
If anyone failed to understand that, it means according to Vasant Shinde, Aryans came from Eastern Europe through Central Asia bringing the ancestral language of Sanskrit to India. He says one thing in the paper and another to the media. A fraud.
Well, there is no contradiction. The so called migration from Eastern Europe via Central Asia is a subsequent development (n the second half of the 2nd millenium BCE, when the Indus Valley civilisation was on the verge of extinction). This woman, whose DNA was analysed lived prior to that. Furthermore, the study says that migration from Iran took place at least 10000 years ago. However, let us not get carried away by analysis of just one sample. This only shows that the Vedic culture was subsequent to the Indus Valley civilisation. Incidentally, Hinduism is still subsequent development being the mixing of vedic philosophy and Buddhism. But there is no mixing of DNA to any great extent. . The concept of Aryan Race is a myth. What we find is just a similarity between European languages and Sanskrit. We all Indians share almost the same DNA.
You seem a little confused. Let me spell it out for you. Modern Indians have up to 30% of their DNA from bronze age steppe migrants, which is totally and utterly missing from the twelve Indus Valley related samples we have available, not to mention dozens more of contemporary samples from Iran and South Central Asia.
So no, the paper is not guessing randomly when they are saying Aryans arrived from Central Asia and mixed with locals in a big way. Every single line of evidence points to it.
My local newspapers are saying Just the opposite of what the study says. They are trying to prove Indo Aryan as being natives and not immigrants.
SHAURYA PRATAP SINGH Sir
I am glad you have realised that you have struck somewhere nearly why some people who are not an anthropologist or a historian or a geneticist who keep on trying to prove which is untrue.. pls read Dr. B.R. Ambedkar’s writings on Indus valley and its decline..
No central asian dna and no Zagros dna either. Why is the print downplaying the Iran dna part of it in the title? Anyway, congratulations for facing up to it, the Caravan and the wire are still reeling from the shock. The findings are in line with textual evidence from the Rigveda and lack of archealogical evidence of central asian migrants.. Shrikant Talageri’s analysis was correct after all.
Modern Indians have 30% central Asian DNA. Pre-collapse IVC people had no central Asian DNA. Shinde thinks Indians are idiots who can’t figure out the implication of these two statements, and you are confirming it.
Finding the supposed Aryan ancestry in IVC would have killed Aryan Invasion Theory, because if the ancestry was already in India, then they couldn’t have migrated again to destroy IVC, could they?
But now AIT stands all but confirmed.
THIS PROVES THE ORIGINAL INHABITANTS OF INDIA WERE NOT ARAYAN AND NOT HINDU THE ARYANS CAME LATER AND BROUGHT THE VEDAS WITH THEM AND INTERMINGLED WITH THE ORIGINAL INHABITANTS OF INDIA. Our own Muslim DNA and the DNA of Hindus has different mixes of DNA, like a pizza which includes ARAYAN and Harrapan. The most aryan DNA is is found in Brahmins this is stark in South India with lower castes.. North Indians both Muslim and Hindus have more ARAYAN DNA than S Indians. Instead of saying “go to Pakistan” to Muslims We can say “go to Central Asia!” To Brahmins. WE ARE ALL INDIANS THE HINDUTVA NARRATIVE ON ANCIENT INDIA IS FALSE
Well shinde and these research cannot prove where “Aryans” came from and what were their beliefs although Aryan INVASION theory is debunked which says that Aryans conquered and imposed hinduism/brahminism on indians , there are various hindu elements in IVC iconography namely pashupatinath seal . Event the Native IVC was a multicultural ( mixed with Iranian people ) which says that native IVC were open to immigrants . Lastly I will say that , carry on with your taqiyya .
Jugglery by Mr.Shindhe and Mrs.Khan.
These are cautious ppl who want to play safe in these days of Big brother’s rule.
They feel safer by putting a convenient foot note of disclaimer without altering the facts.
For the muddle headed Hindutwa morons let me explain:
Absence of steppe genes in those skeletons proves that the so called “Aryans” were not there in Rakhigarhi 5000 years back.
Aryans came much later about 3500-4000 years back. Their genes (steppe gene)are prominently found in present day upper caste north Indians.
For better detailed understanding read Manu Joseph’s book on the issue.
Not to offend anyone, a disclaimer. I know how Manu Joseph went on to write and the connections established thereon. In reality, many such theories are created and concocted stories spun around in the name name of research finding. The very basic purpose of healthy debate is defeating saying Hindutva morons, which itself is like Romila Thaper harping and pushing Nehruvian ideologies as historical research findings and shutting off other historians, bulldozing them in JNU. The very word freedom of expression stops at when Communists get into debate.
The Lost River: On The Trails of Saraswati by Michael Danino has wonderful details of how the Aryan theory was more fictional than actual and the researcher there went on to prove with many examples of aryan invasion is more created than facts. However, Indian hisotorians always have their upper vs lower caste mindset to say, the someone of central asian region invaded and suppressed the rights of locals who are called Drawidians. I am happy that there are more findings unearthed to prove the selective amnesia historians were wrong.
This paper confirms that the Steppe Aryan ancestry in modern Indians was not present in IVC and had to come down from Central Asia… at roughly the time frame traditionally calculated for the Aryan Invasion. Danino can have his fantasies, but facts are not backing him up.
Well, the title is misleading. What Rakhigarhi DNA and genetic mapping say that the Ayyan DNA came after IVC via the Kazakhstan area. So, invasion or not certainly migration post Indus Valley Decline. Migration not from Central asia but from Steppe regions.
Yes 2 groups with highest amount of steppe ancestry are – Brahmins & Bhumihars but the caveat at the end of the paper is necessary to remember that Indus civilization was cosmopolitan & so to base all arguments only on a single sample is misleading & wrong and we all must wait for more samples & results before forming any conclusions.
The eleven outlier individuals DNA is proof enough.
No wonder JNU is asking Romila Thapar her CV. Most of her work is proving to be fake.
U need to learn ABCD from JNU.
This is a (not so)clever conclusion by Mr.Vasanth Sindhe to escape the noose at the hand of Hindutwa big brother. Shindhe is no Galilio. Keeping the facts intact,he puts a disclaimer at the end to save himself. Hindutwa hot heads are entitled to rejoice about nothing.
Romila Thapar is being proven wrong, but not in the way you think. This paper confirms that a large migration of Central Asians happened during or after Indus Valley collapse, and that they provided up to 30% of the DNA of modern Indians.
Thapar was going soft with the ‘pots are not people’ brigade, but they are all being proven wrong these days. Various ancient migrants brought those pots to new lands (and often smashed local peoples’ heads in with them). It wasn’t a few traders who miraculously changed the language of entire nations like Thapar might claim.
I’ve never heard Thapar say that.. she’s always said that Indo Aryans were migrants from the Caucasus region and came from 1900bce to 2600 bce, and that’s what the study has shown.
LEAVE A REPLY Cancel
Save my name, email, and website in this browser for the next time I comment.
Most Popular
Anti-modi groups are a cult now. modern-day ‘shatranj ke khiladi’ playing a losers’ game, ‘was told i got votes due to upar wale log’ — bihar mp ajay nishad, who quit bjp after ticket denial, ‘mongrels, foxes’: ‘injured’ kcr out from pragathi bhavan to tour telangana, slams deserters.
Required fields are marked *
Copyright © 2024 Printline Media Pvt. Ltd. All rights reserved.
- Terms of Use
- Privacy Policy
Civilsdaily
No. 1 UPSC IAS Platform for preparation
Historical and Archaeological Findings in News
Rakhigarhi findings to be mentioned in ncert books.
From UPSC perspective, the following things are important :
Prelims level: DNA Findings of Rakhigarhi, IVC
Mains level: NA

Why in the news?
The National Council of Educational Research and Training (NCERT) has proposed to add information about the DNA analysis of skeletal remains found at Rakhigarhi.

About Rakhigarhi
- The ancient site of Rakhi-Khas and Rakhi-Shahpur are collectively known as Rakhigarhi, located on the right bank of the now dried up Palaeo-channel of Drishadvati .
- It is located in the Ghaggar-Hakra River plain in the Hissar district of Haryana.
- Total seven mounds are located here.
- The site has yielded various stages of Harappan culture and is by far one of the largest Harappan sites in India.
- The site shows the sequential development of the Indus culture in the now dried up Saraswati basin.
Major findings at Rakhi Garhi
- Findings confirm both early and mature Harappan phases and include 4,600-year-old human skeletons , fortification and bricks.
- Digging so far reveals a well-planned city with 92 m wide roads , a bit wider than in Kalibangan.
- The pottery is similar to Kalibangan and Banawali.
- Pits surrounded by walls have been found, which are thought to be for sacrificial or some religious ceremonies.
- There are brick-lined drains to handle sewage from the houses.
- Terracotta statues, weights, bronze artefacts, comb, copper fish hooks , needles and terracotta seals have also been found.
- A bronze vessel has been found which is decorated with gold and silver .
- A granary belonging to the mature Harappan phase has been found here.
- Fire altars structures were revealed in Rakhigarhi.
Get an IAS/IPS ranker as your 1: 1 personal mentor for UPSC 2024

JOIN THE COMMUNITY
Join us across social media platforms., your better version awaits you.
RAKHIGARHI (Syllabus: GS Paper 1 – History)
Context : Recently, NCERT proposed revisions to school textbooks, including additions about DNA analysis findings from Rakhigarhi site and removal of references to Narmada Dam's adverse effects on tribals, aiming for more updated content.
- About : Rakhigarhi stands as a significant site of the ancient Indus Valley Civilization, situated in modern-day Hissar, Haryana.
- Excavation Initiatives by ASI : The Archaeological Survey of India (ASI) initiated the first excavation at Rakhigarhi during 1998-2001.
- Deccan College's Contribution : From 2013 to 2016, Deccan College of Pune conducted excavations at Rakhigarhi, adding to the archaeological understanding of the site.
- Recognition : Rakhigarhi's ancient mounds 6 and 7 have been identified by ASI among 19 sites to be notified as 'sites of national importance.'
- Endangered Status and Global Awareness : In May 2012, Rakhigarhi gained global recognition when the Global Heritage Fund declared it one of the 10 most endangered heritage sites in Asia.
- Cluster of Archaeological Sites : Rakhigarhi is not isolated; it's part of a cluster of significant archaeological sites in the region, including Kalibangan, Kunal, Haryana, Balu, Haryana, Bhirrana, and Banawali, all situated in the old river valley east of the Ghaggar Plain.
Answer our survey to get FREE CONTENT

Feel free to get in touch! We will get back to you shortly
- Privacy Policy
- Terms of Service
- Quality Enrichment Program (QEP)
- Total Enrichment Program (TEP)
- Ethics Marks Maximization Prog. 2024
- Interview Mentorship Program (IMP)
- Prelims Crash Course for UPSC 2024
- Science of Answer Writing (SAW)
- Intensive News Analysis (INA)
- Topper's UPSC PYQ Answer
- Essay Marks Maximization Program
- PSIR Optional
- News-CRUX-10
- Daily Headlines
- Geo. Optional Monthly Editorials
- Past Papers
- © Copyright 2024 - theIAShub
Talk To Our Counsellor

IMAGES
VIDEO
COMMENTS
The findings of a highly anticipated study of ancient DNA from the graveyard of the historic Indian town of Rakhigarhi reveal evidence that will unsettle many Hindutva nationalists. The 'petrous bone' is an inelegant but useful chunk of the human skull -- basically it protects your inner ear. But that's not all it protects.
"There's no doubt this is the most intensive effort we've ever made to get ancient DNA from a single sample," Reich says. The sampled individual, most likely a woman based on her DNA, was buried among dozens of ceramic bowls and vases in an Indus site known as Rakhigarhi, about 150 kilometers northwest of modern-day Delhi.
The scientific paper on the DNA analysis of the Harappan skeleton from Rakhigarhi claims that there was no Aryan invasion and no Aryan migration and that all the developments right from the hunting-gathering stage to modern times in South Asia were done by indigenous people. An agenda-setting for indigeneity of populations, ideas and civilisational traits may be embeded in the paper.
The yellow labels indicate two sites where a minority of buried individuals yielded ancient DNA which matched that of the Rakhigarhi individual. ... the new analysis shows that the first farmers ...
Only a single sample yielded enough authentic ancient DNA for analysis: I6113, Rakhigarhi, Haryana, India (n = 1). We report the archeological context dates for this burial in Method Details. The skeletal samples from Rakhigarhi were excavated by the archaeological team led by V.S. at the Deccan College Post-Graduate and Research Institute in ...
The DNA belongs to a woman who was buried four to five millennia ago in Rakhigarhi, now part of Haryana in India. Her genes point to an ancestry of ancient Iranians and Southeast Asian hunter ...
Dr. Manjul, who is leading the excavation team at the Rakhigarhi site, about 150 km north-west of Delhi, since it commenced on February 24, 2022, said the DNA analysis will help answer a lot of ...
samples from the Rakhigarhi cemetery, which lies 1kmwest of the ancient town (Table S1). We extracted DNA (Dabney et al., ... Population Genetic Analysis (A) PCA of ancient DNA from South and Central Asia projected onto a basis of whole-genome sequencing data from present-day Eurasians. I6113 and I11042
Manjul, who is leading the excavation team at Rakhigarhi site, about 150 km north-west of Delhi, since it commenced on February 24, 2022, said the DNA analysis will help answer a lot of questions ...
Here are some of these findings: 1) No DNA strands either from Iranian or Steppe pastoralists in the Rakhigarhi sample shows that these people were indigenous and they independently developed farming practices in Rakhigarhi. 2) DNA of the Rakhigarhi sample matches with those of modern Bharatiyas. 3) Continuity of culture is visible from ...
DNA analysis might tell about the ancestry and food habits of people who lived in the Rakhigarhi region thousands of years ago. About Rakhi Garhi. The ancient site of Rakhi-Khas and Rakhi-Shahpur are collectively known as Rakhigarhi, located on the right bank of the now dried up Palaeo-channel of Drishadvati.
Rakhigarhi or Rakhi Garhi is a village and an archaeological site belonging to the Indus Valley civilisation in the Hisar District of the northern Indian state of Haryana, ... Paleo-parasitical studies and DNA analysis to determine the lineage is being undertaken.
Manjul, who is leading the excavation team at Rakhigarhi site, about 150 km north-west of Delhi, since it commenced on February 24, 2022, said the DNA analysis will help answer a lot of questions ...
The DNA sample retrieved from Rakhigarhi gave an impetus to a long-drawn debate and controversy. This sample was extracted from one of over 60 burials excavated at RGR 7 located about 500 m north of the lapidary centre, (RGR 1) in the agricultural field. Buried in privately owned agriculture fields, the remains of this 'cemetery' of the ...
A woman's skeleton from 2500 BC has finally spoken. A team of Indian and international scientists and archaeologists — who conducted a genetic analysis of the remains of a woman buried in Rakhigarhi, a Harappan site in Haryana — have found no traces of the R1a1 gene, which is often loosely called the 'Aryan gene'. This new finding has set off a debate about history, politics and ...
Rakhigarhi (Har), May 8 (PTI) DNA samples collected from two human skeletons unearthed at a necropolis of a Harappan-era city site in Haryana have been sent for scientific examination, the outcome of which might tell about the ancestry and food habits of people who lived in Rakhigarhi region thousands of years ago.
Only a single sample yielded enough authentic ancient DNA for analysis: I6113, Rakhigarhi, Haryana, India (n=1). We report the archeological context dates for this burial in Method Details. The skeletal samples from Rakhigarhi were excavated by the archaeological team led by V.S. at the Deccan College Post-Graduate and Research Institute in ...
Rakhigarhi (Har), May 8 (PTI) DNA samples collected from two human skeletons unearthed at a necropolis of a Harappan-era city site in Haryana have been sent for scientific examination, the outcome of which might tell about the ancestry and food habits of people who lived in Rakhigarhi region thousands of years ago. The skeletons of two women were found a couple of months ago at mound number 7 ...
Manjul, who is leading the excavation team at Rakhigarhi site, about 150 km north-west of Delhi, since it commenced on February 24, 2022, said the DNA analysis will help answer a lot of questions ...
The study argues that the DNA analysis shows that the people at Rakhigarhi, and the individual whose skeleton was examined, are from a population that is the largest source of ancestry for south ...
The Cell paper shows that the single skeleton that yielded ancient DNA was of a woman who was ceremonially buried roughly about 4,500-5,000 years ago. This would mean it predated the steppes migration into South Asia which takes place at least 600-1000 years after that. The Rakhigarhi ancient DNA shows the absence of any steppes signature, and ...
A-. A+. New Delhi: The study of DNA samples of the skeletons found in Rakhigarhi, an Indus Valley Civilisation site in Haryana, has found no traces of the R1a1 gene or Central Asian 'steppe' genes, loosely termed as the 'Aryan gene'. The study — titled 'An ancient Harappan genome lacks ancestry from Steppe pastoralists or Iranian ...
The analysis of the data was interpreted by some that the Harappans are the indigenous people of this region. The same team of researchers also reconstructed 3D features of the Harappan people.
DNA Findings of Rakhigarhi. This research was conducted by the Deccan College Deemed University, Pune, in collaboration with the Centre for Cellular and Molecular Biology, Hyderabad, and The Harvard Medical College.. Their key findings include-Debunking of the Aryan Invasion Theory.; The genetic roots of the Harappans date back to 10,000 BCE.; DNA of the Harappans has continued to exist among ...
Study of DNA Samples: Recently, a study of DNA from skeletal remains excavated from the Harappan cemetery at Rakhigarhi found that the people in the Harappan Civilization have an independent origin. This study negates the theory of the Harappans having Steppe pastoral or ancient Iranian farmer ancestry.
Catalog; For You; The Hindu - International. NCERT likely to add Rakhigarhi findings in books The National Council of Educational Research and Training (NCERT) has proposed that findings from the DNA analysis of skeletal remains unearthed at the archaeological site of Rakhigarhi in Haryana be added to the Class 12 history textbook.
Context: Recently, NCERT proposed revisions to school textbooks, including additions about DNA analysis findings from Rakhigarhi site and removal of references to Narmada Dam's adverse effects on tribals, aiming for more updated content.. Rakhigarhi. About: Rakhigarhi stands as a significant site of the ancient Indus Valley Civilization, situated in modern-day Hissar, Haryana.
DNA analysis of unmarked remains has shed light on a long-standing mystery surrounding the fates of President George Washington's younger brother Samuel and his kin.
Officials using DNA analysis have identified the remains of a 19-year-old Virginian sailor killed in a World War II attack more than 80 years ago, the United States Defense POW/MIA Accounting ...